import numpy as np
import pandas as pd
import matplotlib
import matplotlib.pyplot as plt
from sklearn.preprocessing import MinMaxScaler
from sklearn.metrics import mean_absolute_error, mean_squared_error
from keras import Input
from keras.models import Sequential
from keras.layers import Dense, LSTM
from keras.layers.core import Dense, Activation, DropoutLSTM univariate prediction
Import python packages:
data_covid = pd.read_csv('data/clean/final_covid_data.csv')
data_covid| provincia | fecha | num_casos | num_casos_prueba_pcr | num_casos_prueba_test_ac | num_casos_prueba_ag | num_casos_prueba_elisa | num_casos_prueba_desconocida | num_hosp | num_uci | ... | ws | ws_max | sol | mob_grocery_pharmacy | mob_parks | mob_residential | mob_retail_recreation | mob_transit_stations | mob_workplaces | mob_flujo | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Barcelona | 2020-01-01 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 2.5 | 7.2 | 4.9 | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 1 | Madrid | 2020-01-01 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | ... | 0.8 | 3.6 | 8.3 | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 2 | Málaga | 2020-01-01 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 3.3 | 6.7 | 7.7 | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 3 | Asturias | 2020-01-01 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 2.5 | 7.8 | 7.9 | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 4 | Sevilla | 2020-01-01 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | ... | 1.9 | 5.8 | 9.1 | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 4090 | Barcelona | 2022-03-29 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 7.2 | 13.3 | 0.0 | 0.0 | -13.0 | 5.0 | -24.0 | -16.0 | -17.0 | NaN |
| 4091 | Madrid | 2022-03-29 | 6 | 3 | 0 | 3 | 0 | 0 | 0 | 0 | ... | 2.2 | 6.1 | 2.4 | 1.0 | -11.0 | 4.0 | -25.0 | -16.0 | -16.0 | NaN |
| 4092 | Málaga | 2022-03-29 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 4.2 | 10.8 | 1.5 | 4.0 | -3.0 | 4.0 | -16.0 | 1.0 | -8.0 | NaN |
| 4093 | Asturias | 2022-03-29 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | ... | 2.2 | 6.7 | 0.0 | -4.0 | 17.0 | 3.0 | -25.0 | -15.0 | -12.0 | NaN |
| 4094 | Sevilla | 2022-03-29 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 2.5 | 8.3 | 1.6 | 3.0 | -7.0 | 2.0 | -15.0 | -13.0 | -5.0 | NaN |
4095 rows × 26 columns
All the available data
We will only detele data from the first wave since it is not reliable.
Asturias
data_asturias = data_covid.loc[data_covid['provincia'] == 'Asturias']
data_asturias = data_asturias.set_index('fecha')
data_asturias = data_asturias.filter(['num_casos'])
data_asturias = data_asturias['2020-06-14':]
data_asturias| num_casos | |
|---|---|
| fecha | |
| 2020-06-14 | 0 |
| 2020-06-15 | 0 |
| 2020-06-16 | 1 |
| 2020-06-17 | 0 |
| 2020-06-18 | 0 |
| ... | ... |
| 2022-03-25 | 244 |
| 2022-03-26 | 432 |
| 2022-03-27 | 1 |
| 2022-03-28 | 9 |
| 2022-03-29 | 0 |
654 rows × 1 columns
data_asturias.describe()| num_casos | |
|---|---|
| count | 654.00000 |
| mean | 312.11315 |
| std | 565.17985 |
| min | 0.00000 |
| 25% | 43.50000 |
| 50% | 117.00000 |
| 75% | 323.75000 |
| max | 3827.00000 |
np_data_asturias = data_asturias.valuesscaler = MinMaxScaler(feature_range=(0, 1))
scaled_data_asturias = scaler.fit_transform(np_data_asturias)
print(f'Longitud del conjunto de datos disponible: {len(scaled_data_asturias)}')Longitud del conjunto de datos disponible: 654# Since we are going to predict future values based on the X past elements,
# we need to create a list with those historic information for each element
historic_values = 90
scaled_data_asturias_x = []
scaled_data_asturias_y = []
for num_casos_i in range(historic_values, len(scaled_data_asturias)):
scaled_data_asturias_x.append(scaled_data_asturias[(num_casos_i-historic_values):num_casos_i, 0])
scaled_data_asturias_y.append(scaled_data_asturias[num_casos_i, 0])
# Convert the x_train and y_train to numpy arrays
scaled_data_asturias_x = np.array(scaled_data_asturias_x)
scaled_data_asturias_y = np.array(scaled_data_asturias_y)# Train data looks like
scaled_data_asturias_x[235]array([0.07002874, 0.0611445 , 0.07107395, 0.07394826, 0.06689313,
0.05644108, 0.05278286, 0.03841129, 0.03501437, 0.04285341,
0.04990854, 0.05016985, 0.04285341, 0.03710478, 0.03057225,
0.03240136, 0.03945649, 0.03971779, 0.03762738, 0.04311471,
0.02926574, 0.02691403, 0.03945649, 0.03919519, 0.03396917,
0.04206951, 0.03684348, 0.03841129, 0.03475307, 0.02795924,
0.02560753, 0.02743663, 0.03788869, 0.03814999, 0.02769794,
0.03031095, 0.02822054, 0.03893389, 0.03240136, 0.03553697,
0.03971779, 0.01802979, 0.01646198, 0.03004965, 0.03109485,
0.02926574, 0.04468252, 0.03266266, 0.02482362, 0.03031095,
0.03109485, 0.03893389, 0.03579828, 0.02325581, 0.03841129,
0.0195976 , 0.02508492, 0.02456232, 0.03057225, 0.03527567,
0.03919519, 0.03605958, 0.02534622, 0.02926574, 0.03240136,
0.04807944, 0.0399791 , 0.03396917, 0.02743663, 0.02325581,
0.02613013, 0.03083355, 0.03788869, 0.02848184, 0.03344656,
0.03396917, 0.02822054, 0.02247191, 0.02351712, 0.02586883,
0.02534622, 0.02874314, 0.0198589 , 0.0211654 , 0.01254246,
0.01907499, 0.01228116, 0.01228116, 0.01881369, 0.01515547])# Test data looks like
scaled_data_asturias_y[235]0.013326365299189964# Since the first 90 values does not have historic, the dataset has been reduced in 90 values
print(f'Longitud datos de entrenamiento con historico: {len(scaled_data_asturias_y)}')Longitud datos de entrenamiento con historico: 564# we split data in train and test
# as in previous analysis, we are going to predict a maximum of 21 days
x_train = scaled_data_asturias_x[0:len(scaled_data_asturias_x)-91]
y_train = scaled_data_asturias_y[0:len(scaled_data_asturias_y)-91]
print(f'Cantidad datos de entrenamiento: x={len(x_train)} - y={len(y_train)}')
x_test = scaled_data_asturias_x[len(scaled_data_asturias_x)-90:len(scaled_data_asturias_x)]
y_test = scaled_data_asturias_y[len(scaled_data_asturias_y)-90:len(scaled_data_asturias_y)]
print(f'Cantidad datos de test: x={len(x_test)} - y={len(y_test)}')Cantidad datos de entrenamiento: x=473 - y=473
Cantidad datos de test: x=90 - y=90# Reshape the data to feed de recurrent network
x_train = np.reshape(x_train, (x_train.shape[0], x_train.shape[1], 1))
print("Train data shape:")
print(x_train.shape)
print(y_train.shape)
x_test = np.reshape(x_test, (x_test.shape[0], x_test.shape[1], 1))
print("Test data shape:")
print(x_test.shape)
print(y_test.shape)Train data shape:
(473, 90, 1)
(473,)
Test data shape:
(90, 90, 1)
(90,)# # Configure / setup the neural network model - LSTM
# Build the model
print('Build model...')
model = Sequential()
# Model with Neurons
# Inputshape = neurons -> Timestamps
neurons= x_train.shape[1]
model.add(LSTM(90,
activation = 'relu',
return_sequences = True,
input_shape = (x_train.shape[1], 1)))
model.add(LSTM(50,
activation = 'relu',
return_sequences = True))
model.add(LSTM(25,
activation = 'relu',
return_sequences = False))
model.add(Dense(5, activation = 'relu'))
model.add(Dense(1))Build model...2022-05-25 01:22:54.266575: I tensorflow/core/platform/cpu_feature_guard.cc:193] This TensorFlow binary is optimized with oneAPI Deep Neural Network Library (oneDNN) to use the following CPU instructions in performance-critical operations: AVX2 FMA
To enable them in other operations, rebuild TensorFlow with the appropriate compiler flags.model.compile(optimizer='adam', loss='mean_squared_error')
model.summary()Model: "sequential"_________________________________________________________________ Layer (type) Output Shape Param # ================================================================= lstm (LSTM) (None, 90, 90) 33120 lstm_1 (LSTM) (None, 90, 50) 28200 lstm_2 (LSTM) (None, 25) 7600 dense (Dense) (None, 5) 130 dense_1 (Dense) (None, 1) 6 =================================================================Total params: 69,056Trainable params: 69,056Non-trainable params: 0_________________________________________________________________# Training the model
# # fit network
history = model.fit(x_train,
y_train,
batch_size=1000,
epochs = 30,
validation_data = (x_test, y_test),
verbose = 0)plt.plot(history.history['loss'], label='train')
plt.plot(history.history['val_loss'], label='test')
plt.legend()
plt.show() 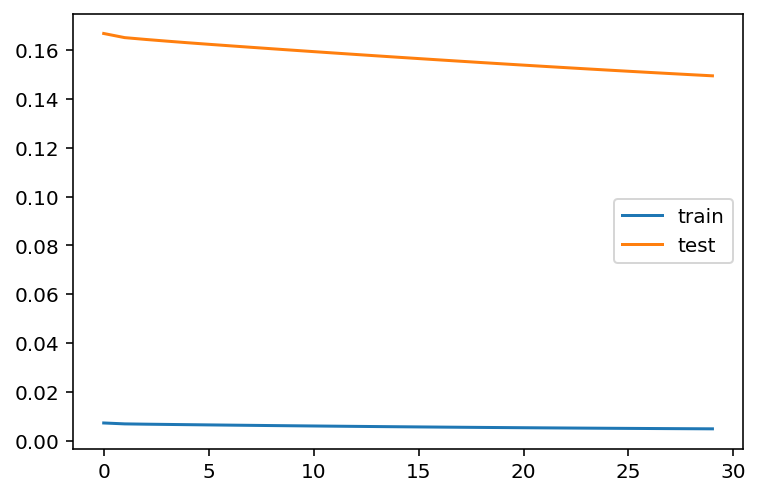
# Get the predicted values
predictions = model.predict(x_test)
predictions = scaler.inverse_transform(predictions)1/3 [=========>....................] - ETA: 0s3/3 [==============================] - ETA: 0s3/3 [==============================] - 0s 31ms/stepy_test = y_test.reshape(-1,1)
y_test = scaler.inverse_transform(y_test)# Calculate the mean absolute error (MAE)
mae = mean_absolute_error(y_test, predictions)
print('MAE: ' + str(round(mae, 1)))
# Calculate the root mean squarred error (RMSE)
rmse = np.sqrt(mean_squared_error(y_test,predictions))
print('RMSE: ' + str(round(rmse, 1)))
# Calculate the root mean squarred error (RMSE)
rmse = mean_squared_error(y_test,
predictions,
squared = False)
print('RMSE: ' + str(round(rmse, 1)))MAE: 1074.0
RMSE: 1479.1
RMSE: 1479.1# Add the difference between the valid and predicted prices
train = data_asturias[:(len(x_train)+92)]
valid = data_asturias[(len(x_train)+91):]valid.insert(1, "Predictions", predictions, True)
valid.insert(1, "Difference", valid["Predictions"] - valid["num_casos"], True)# Zoom-in to a closer timeframe
# Date from which on the date is displayed
display_start_date = "2021-10-31"
valid = valid[valid.index > display_start_date]
train = train[train.index > display_start_date]# Visualize the data
matplotlib.style.use('ggplot')
fig, ax1 = plt.subplots(figsize=(22, 10), sharex=True)
# Data - Train
xt = train.index;
yt = train[["num_casos"]]
# Data - Test / validation
xv = valid.index;
yv = valid[["num_casos", "Predictions"]]
# Plot
plt.title("Asturias: Predictions vs Real infections", fontsize=20)
plt.ylabel("Nº Cases", fontsize=18)
plt.plot(yt, color="blue", linewidth=1.5)
plt.plot(yv["Predictions"], color="red", linewidth=1.5)
plt.plot(yv["num_casos"], color="green", linewidth=1.5)
plt.legend(["Train", "LSTM Predictions", "Test"],
loc="upper left", fontsize=18)
# Bar plot with the differences
x = valid.index
y = valid["Difference"]
plt.bar(x, y, width=0.2, color="grey")
plt.grid()
plt.show()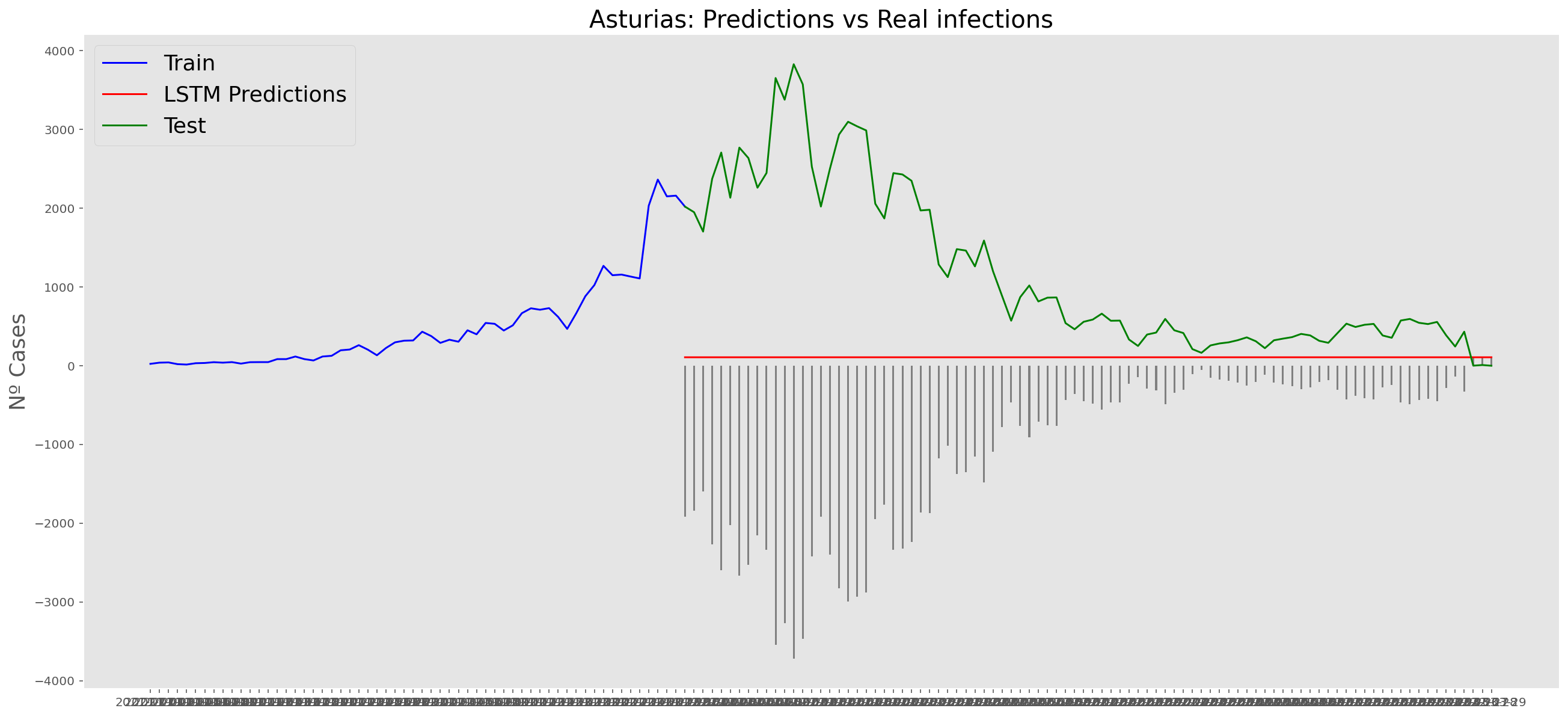
Barcelona
data_Barcelona = data_covid.loc[data_covid['provincia'] == 'Barcelona']
data_Barcelona = data_Barcelona.set_index('fecha')
data_Barcelona = data_Barcelona.filter(['num_casos'])
data_Barcelona = data_Barcelona['2020-06-14':]
data_Barcelona| num_casos | |
|---|---|
| fecha | |
| 2020-06-14 | 33 |
| 2020-06-15 | 62 |
| 2020-06-16 | 66 |
| 2020-06-17 | 70 |
| 2020-06-18 | 68 |
| ... | ... |
| 2022-03-25 | 598 |
| 2022-03-26 | 345 |
| 2022-03-27 | 252 |
| 2022-03-28 | 688 |
| 2022-03-29 | 0 |
654 rows × 1 columns
data_Barcelona.describe()| num_casos | |
|---|---|
| count | 654.000000 |
| mean | 2605.048930 |
| std | 4823.001644 |
| min | 0.000000 |
| 25% | 597.250000 |
| 50% | 1030.500000 |
| 75% | 2243.750000 |
| max | 34701.000000 |
np_data_Barcelona = data_Barcelona.valuesscaler = MinMaxScaler(feature_range=(0, 1))
scaled_data_Barcelona = scaler.fit_transform(np_data_Barcelona)
print(f'Longitud del conjunto de datos disponible: {len(scaled_data_Barcelona)}')Longitud del conjunto de datos disponible: 654# Since we are going to predict future values based on the 90 past elements,
# we need to create a list with those historic information for each element
historic_values = 90
scaled_data_Barcelona_x = []
scaled_data_Barcelona_y = []
for num_casos_i in range(historic_values, len(scaled_data_Barcelona)):
scaled_data_Barcelona_x.append(scaled_data_Barcelona[(num_casos_i-historic_values):num_casos_i, 0])
scaled_data_Barcelona_y.append(scaled_data_Barcelona[num_casos_i, 0])
# Convert the x_train and y_train to numpy arrays
scaled_data_Barcelona_x = np.array(scaled_data_Barcelona_x)
scaled_data_Barcelona_y = np.array(scaled_data_Barcelona_y)# Train data looks like
scaled_data_Barcelona_x[235]array([0.05037319, 0.04726089, 0.02899052, 0.02507132, 0.04639636,
0.04190081, 0.03749171, 0.03527276, 0.03786634, 0.02619521,
0.02351517, 0.04077692, 0.03760699, 0.03587793, 0.0364831 ,
0.03120948, 0.02449497, 0.02083513, 0.03792398, 0.03613729,
0.03195873, 0.02754964, 0.02818363, 0.02025878, 0.01752111,
0.03484049, 0.03037376, 0.02870234, 0.0233999 , 0.02610876,
0.01876027, 0.01645486, 0.0282989 , 0.02625285, 0.02680038,
0.02564768, 0.02596467, 0.01953834, 0.01746347, 0.03184346,
0.02973978, 0.02573413, 0.02838535, 0.02919224, 0.02190139,
0.01890435, 0.03720354, 0.03642546, 0.0325639 , 0.0304314 ,
0.03556093, 0.02524423, 0.02149794, 0.03803925, 0.03426414,
0.03048903, 0.03365897, 0.02218956, 0.02530186, 0.02533068,
0.02832771, 0.03835624, 0.03740526, 0.03512867, 0.03680009,
0.02855825, 0.02118095, 0.03878851, 0.03507104, 0.03985476,
0.03155529, 0.03394715, 0.02423561, 0.02169966, 0.03953777,
0.04596409, 0.0350134 , 0.0395954 , 0.03267917, 0.02772254,
0.02380335, 0.04668453, 0.04213135, 0.03806807, 0.03671364,
0.03509985, 0.02106568, 0.01694476, 0.03803925, 0.02815481])# Test data looks like
scaled_data_Barcelona_y[235]0.030661940578081325# Since the first 90th values does not have historic, the dataset has been reduced in 90 values
print(f'Longitud datos de entrenamiento con historico: {len(scaled_data_Barcelona_y)}')Longitud datos de entrenamiento con historico: 564# we split data in train and test
# as in previous analysis, we are going to predict a maximum of 90 days
x_train = scaled_data_Barcelona_x[0:len(scaled_data_Barcelona_x)-91]
y_train = scaled_data_Barcelona_y[0:len(scaled_data_Barcelona_y)-91]
print(f'Cantidad datos de entrenamiento: x={len(x_train)} - y={len(y_train)}')
x_test = scaled_data_Barcelona_x[len(scaled_data_Barcelona_x)-90:len(scaled_data_Barcelona_x)]
y_test = scaled_data_Barcelona_y[len(scaled_data_Barcelona_y)-90:len(scaled_data_Barcelona_y)]
print(f'Cantidad datos de test: x={len(x_test)} - y={len(y_test)}')Cantidad datos de entrenamiento: x=473 - y=473
Cantidad datos de test: x=90 - y=90# Reshape the data to feed de recurrent network
x_train = np.reshape(x_train, (x_train.shape[0], x_train.shape[1], 1))
print("Train data shape:")
print(x_train.shape)
print(y_train.shape)
x_test = np.reshape(x_test, (x_test.shape[0], x_test.shape[1], 1))
print("Test data shape:")
print(x_test.shape)
print(y_test.shape)Train data shape:
(473, 90, 1)
(473,)
Test data shape:
(90, 90, 1)
(90,)# Configure / setup the neural network model - LSTM
# Build the model
print('Build model...')
model = Sequential()
# Model with Neurons
# Inputshape = neurons -> Timestamps
neurons= x_train.shape[1]
model.add(LSTM(90,
activation = 'relu',
return_sequences = True,
input_shape = (x_train.shape[1], 1)))
model.add(LSTM(50,
activation = 'relu',
return_sequences = True))
model.add(LSTM(25,
activation = 'relu',
return_sequences = False))
model.add(Dense(5, activation = 'relu'))
model.add(Dense(1))Build model...model.compile(optimizer='adam', loss='mean_squared_error')
model.summary()Model: "sequential_1"_________________________________________________________________ Layer (type) Output Shape Param # ================================================================= lstm_3 (LSTM) (None, 90, 90) 33120 lstm_4 (LSTM) (None, 90, 50) 28200 lstm_5 (LSTM) (None, 25) 7600 dense_2 (Dense) (None, 5) 130 dense_3 (Dense) (None, 1) 6 =================================================================Total params: 69,056Trainable params: 69,056Non-trainable params: 0_________________________________________________________________# Training the model
# fit network
history = model.fit(x_train,
y_train,
batch_size = 1000,
epochs = 30,
validation_data = (x_test, y_test),
verbose = 0)plt.plot(history.history['loss'], label='train')
plt.plot(history.history['val_loss'], label='test')
plt.legend()
plt.show() 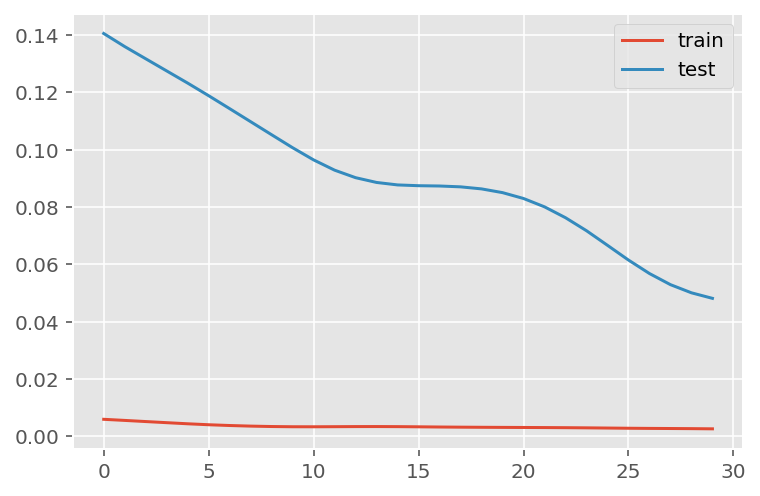
# Get the predicted values
predictions = model.predict(x_test)
predictions = scaler.inverse_transform(predictions)1/3 [=========>....................] - ETA: 0s3/3 [==============================] - ETA: 0s3/3 [==============================] - 0s 31ms/stepy_test = y_test.reshape(-1,1)
y_test = scaler.inverse_transform(y_test)# Calculate the mean absolute error (MAE)
mae = mean_absolute_error(y_test, predictions)
print('MAE: ' + str(round(mae, 1)))
# Calculate the root mean squarred error (RMSE)
rmse = np.sqrt(mean_squared_error(y_test,predictions))
print('RMSE: ' + str(round(rmse, 1)))
# Calculate the root mean squarred error (RMSE)
rmse = mean_squared_error(y_test,
predictions,
squared = False)
print('RMSE: ' + str(round(rmse, 1)))MAE: 5228.7
RMSE: 7609.8
RMSE: 7609.8# Add the difference between the valid and predicted prices
train = data_Barcelona[:(len(x_train)+92)]
valid = data_Barcelona[(len(x_train)+91):]valid.insert(1, "Predictions", predictions, True)
valid.insert(1, "Difference", valid["Predictions"] - valid["num_casos"], True)# Zoom-in to a closer timeframe
# Date from which on the date is displayed
display_start_date = "2021-10-31"
valid = valid[valid.index > display_start_date]
train = train[train.index > display_start_date]# Visualize the data
matplotlib.style.use('ggplot')
fig, ax1 = plt.subplots(figsize=(22, 10), sharex=True)
# Data - Train
xt = train.index;
yt = train[["num_casos"]]
# Data - Test / validation
xv = valid.index;
yv = valid[["num_casos", "Predictions"]]
# Plot
plt.title("Barcelona: Predictions vs Real infections", fontsize=20)
plt.ylabel("Nº Cases", fontsize=18)
plt.plot(yt, color="blue", linewidth=1.5)
plt.plot(yv["Predictions"], color="red", linewidth=1.5)
plt.plot(yv["num_casos"], color="green", linewidth=1.5)
plt.legend(["Train", "LSTM Predictions", "Test"],
loc="upper left", fontsize=18)
# Bar plot with the differences
x = valid.index
y = valid["Difference"]
plt.bar(x, y, width=0.2, color="grey")
plt.grid()
plt.show()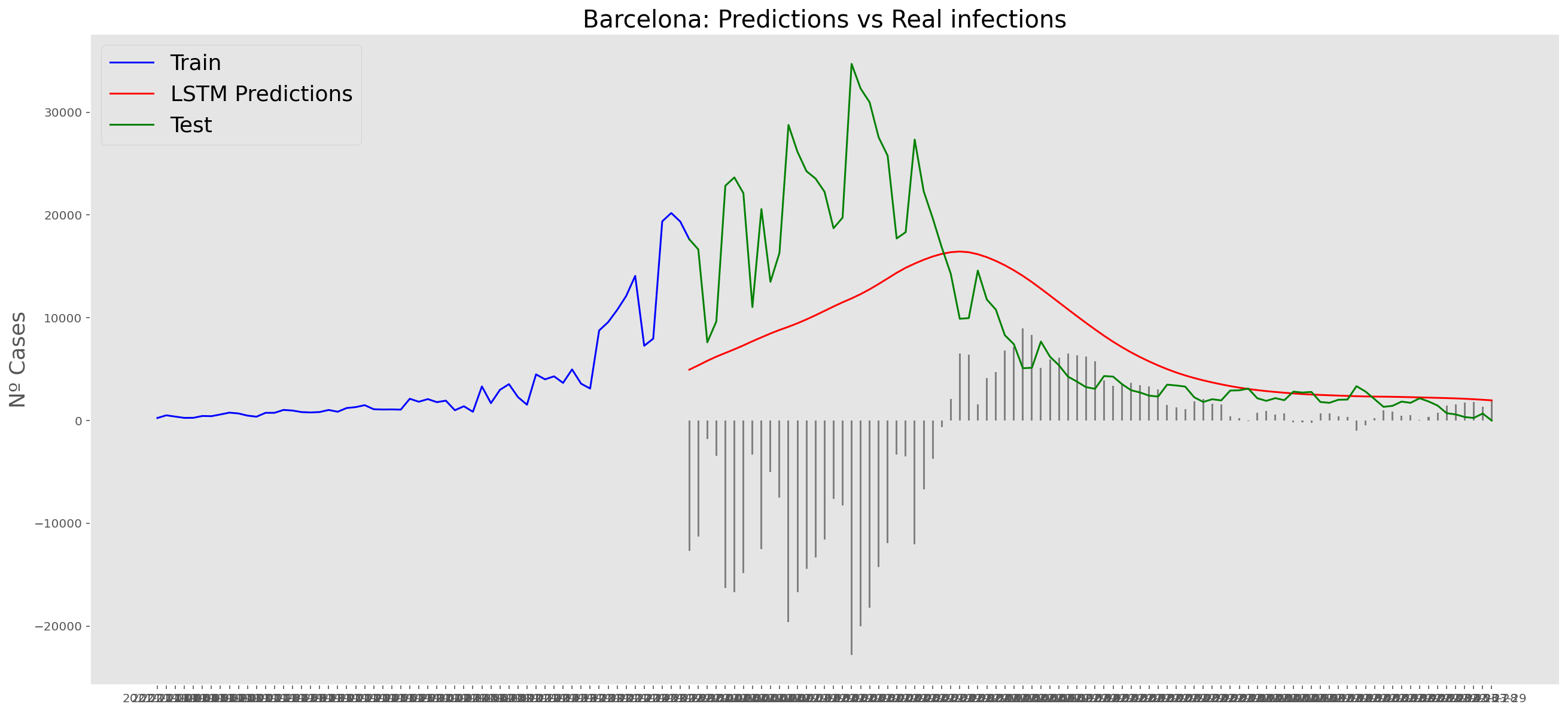
Madrid
data_Madrid = data_covid.loc[data_covid['provincia'] == 'Madrid']
data_Madrid = data_Madrid.set_index('fecha')
data_Madrid = data_Madrid.filter(['num_casos'])
data_Madrid = data_Madrid['2020-06-14':]
data_Madrid| num_casos | |
|---|---|
| fecha | |
| 2020-06-14 | 81 |
| 2020-06-15 | 153 |
| 2020-06-16 | 91 |
| 2020-06-17 | 93 |
| 2020-06-18 | 85 |
| ... | ... |
| 2022-03-25 | 356 |
| 2022-03-26 | 303 |
| 2022-03-27 | 77 |
| 2022-03-28 | 839 |
| 2022-03-29 | 6 |
654 rows × 1 columns
data_Madrid.describe()| num_casos | |
|---|---|
| count | 654.000000 |
| mean | 2392.562691 |
| std | 3390.272836 |
| min | 6.000000 |
| 25% | 662.750000 |
| 50% | 1413.000000 |
| 75% | 2475.750000 |
| max | 23811.000000 |
np_data_Madrid = data_Madrid.valuesscaler = MinMaxScaler(feature_range=(0, 1))
scaled_data_Madrid = scaler.fit_transform(np_data_Madrid)
print(f'Longitud del conjunto de datos disponible: {len(scaled_data_Madrid)}')Longitud del conjunto de datos disponible: 654# Since we are going to predict future values based on the X past elements,
# we need to create a list with those historic information for each element
historic_values = 90
scaled_data_Madrid_x = []
scaled_data_Madrid_y = []
for num_casos_i in range(historic_values, len(scaled_data_Madrid)):
scaled_data_Madrid_x.append(scaled_data_Madrid[(num_casos_i-historic_values):num_casos_i, 0])
scaled_data_Madrid_y.append(scaled_data_Madrid[num_casos_i, 0])
# Convert the x_train and y_train to numpy arrays
scaled_data_Madrid_x = np.array(scaled_data_Madrid_x)
scaled_data_Madrid_y = np.array(scaled_data_Madrid_y)# Train data looks like
scaled_data_Madrid_x[235]array([0.11510187, 0.14820416, 0.08880487, 0.06481832, 0.11169922,
0.11354757, 0.089477 , 0.0694812 , 0.09300567, 0.06133165,
0.04709095, 0.08447805, 0.07746272, 0.06595253, 0.05708885,
0.06515438, 0.04835119, 0.04377232, 0.06372611, 0.05931527,
0.05141777, 0.04541063, 0.05330813, 0.04150389, 0.04032766,
0.05486242, 0.05225793, 0.04822516, 0.04259609, 0.05246797,
0.03919345, 0.03860534, 0.05561857, 0.05452636, 0.04814115,
0.04448645, 0.05288805, 0.04125184, 0.03994959, 0.05448435,
0.05183785, 0.0531401 , 0.05713085, 0.0431842 , 0.05061962,
0.0478891 , 0.07292586, 0.07830288, 0.0626339 , 0.05902121,
0.07485822, 0.05326612, 0.05335014, 0.07111951, 0.07095148,
0.07784079, 0.05469439, 0.06166772, 0.07149758, 0.07456417,
0.09880277, 0.09850872, 0.0857803 , 0.08178954, 0.08951901,
0.07225373, 0.05977736, 0.09750053, 0.09762655, 0.08930897,
0.08544423, 0.09489603, 0.07187566, 0.06406217, 0.09329973,
0.09855072, 0.08435203, 0.07653854, 0.08905692, 0.0647343 ,
0.05486242, 0.08237765, 0.0873766 , 0.07326192, 0.06259189,
0.07485822, 0.04654484, 0.04494854, 0.04721697, 0.06700273])# Test data looks like
scaled_data_Madrid_y[235]0.0648183154799412# Since the first 90th values does not have historic, the dataset has been reduced in 90 values
print(f'Longitud datos de entrenamiento con historico: {len(scaled_data_Madrid_y)}')Longitud datos de entrenamiento con historico: 564# we split data in train and test
# as in previous analysis, we are going to predict a maximum of 91 days
x_train = scaled_data_Madrid_x[0:len(scaled_data_Madrid_x)-91]
y_train = scaled_data_Madrid_y[0:len(scaled_data_Madrid_y)-91]
print(f'Cantidad datos de entrenamiento: x={len(x_train)} - y={len(y_train)}')
x_test = scaled_data_Madrid_x[len(scaled_data_Madrid_x)-90:len(scaled_data_Madrid_x)]
y_test = scaled_data_Madrid_y[len(scaled_data_Madrid_y)-90:len(scaled_data_Madrid_y)]
print(f'Cantidad datos de test: x={len(x_test)} - y={len(y_test)}')Cantidad datos de entrenamiento: x=473 - y=473
Cantidad datos de test: x=90 - y=90# Reshape the data to feed de recurrent network
x_train = np.reshape(x_train, (x_train.shape[0], x_train.shape[1], 1))
print("Train data shape:")
print(x_train.shape)
print(y_train.shape)
x_test = np.reshape(x_test, (x_test.shape[0], x_test.shape[1], 1))
print("Test data shape:")
print(x_test.shape)
print(y_test.shape)Train data shape:
(473, 90, 1)
(473,)
Test data shape:
(90, 90, 1)
(90,)# Configure / setup the neural network model - LSTM
# Build the model
print('Build model...')
model = Sequential()
# Model with Neurons
# Inputshape = neurons -> Timestamps
neurons= x_train.shape[1]
model.add(LSTM(90,
activation = 'relu',
return_sequences = True,
input_shape = (x_train.shape[1], 1)))
model.add(LSTM(50,
activation = 'relu',
return_sequences = True))
model.add(LSTM(25,
activation = 'relu',
return_sequences = False))
model.add(Dense(5, activation = 'relu'))
model.add(Dense(1))Build model...model.compile(optimizer='adam', loss='mean_squared_error')
model.summary()Model: "sequential_2"_________________________________________________________________ Layer (type) Output Shape Param # ================================================================= lstm_6 (LSTM) (None, 90, 90) 33120 lstm_7 (LSTM) (None, 90, 50) 28200 lstm_8 (LSTM) (None, 25) 7600 dense_4 (Dense) (None, 5) 130 dense_5 (Dense) (None, 1) 6 =================================================================Total params: 69,056Trainable params: 69,056Non-trainable params: 0_________________________________________________________________# Training the model
# fit network
history = model.fit(x_train,
y_train,
batch_size = 1000,
epochs = 30,
validation_data = (x_test, y_test),
verbose = 0)plt.plot(history.history['loss'], label='train')
plt.plot(history.history['val_loss'], label='test')
plt.legend()
plt.show() 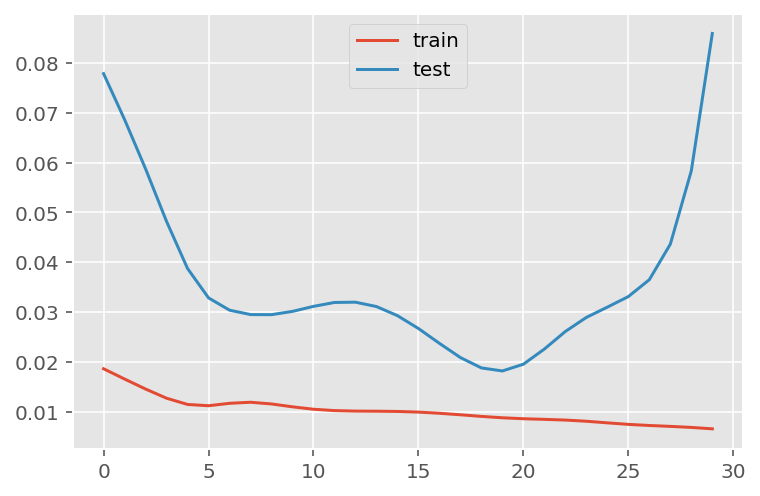
# Get the predicted values
predictions = model.predict(x_test)
predictions = scaler.inverse_transform(predictions)1/3 [=========>....................] - ETA: 0s3/3 [==============================] - ETA: 0s3/3 [==============================] - 0s 30ms/stepy_test = y_test.reshape(-1,1)
y_test = scaler.inverse_transform(y_test)# Calculate the mean absolute error (MAE)
mae = mean_absolute_error(y_test, predictions)
print('MAE: ' + str(round(mae, 1)))
# Calculate the root mean squarred error (RMSE)
rmse = np.sqrt(mean_squared_error(y_test,predictions))
print('RMSE: ' + str(round(rmse, 1)))
# Calculate the root mean squarred error (RMSE)
rmse = mean_squared_error(y_test,
predictions,
squared = False)
print('RMSE: ' + str(round(rmse, 1)))MAE: 4748.2
RMSE: 6975.8
RMSE: 6975.8# Add the difference between the valid and predicted prices
train = data_Madrid[:(len(x_train)+92)]
valid = data_Madrid[(len(x_train)+91):]valid.insert(1, "Predictions", predictions, True)
valid.insert(1, "Difference", valid["Predictions"] - valid["num_casos"], True)# Zoom-in to a closer timeframe
# Date from which on the date is displayed
display_start_date = "2021-10-31"
valid = valid[valid.index > display_start_date]
train = train[train.index > display_start_date]# Visualize the data
matplotlib.style.use('ggplot')
fig, ax1 = plt.subplots(figsize=(22, 10), sharex=True)
# Data - Train
xt = train.index;
yt = train[["num_casos"]]
# Data - Test / validation
xv = valid.index;
yv = valid[["num_casos", "Predictions"]]
# Plot
plt.title("Madrid: Predictions vs Real infections", fontsize=20)
plt.ylabel("Nº Cases", fontsize=18)
plt.plot(yt, color="blue", linewidth=1.5)
plt.plot(yv["Predictions"], color="red", linewidth=1.5)
plt.plot(yv["num_casos"], color="green", linewidth=1.5)
plt.legend(["Train", "LSTM Predictions", "Test"],
loc="upper left", fontsize=18)
# Bar plot with the differences
x = valid.index
y = valid["Difference"]
plt.bar(x, y, width=0.2, color="grey")
plt.grid()
plt.show()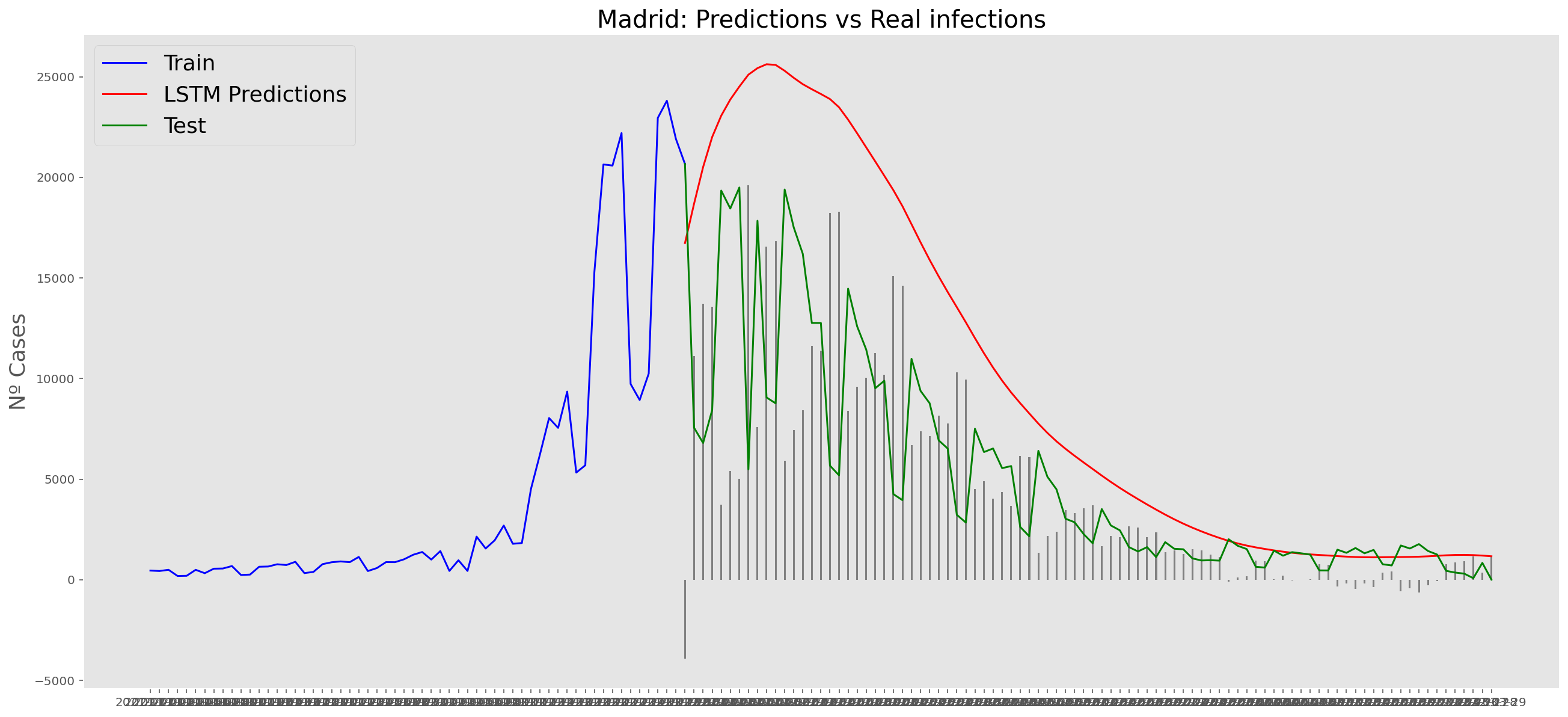
Malaga
data_Malaga = data_covid.loc[data_covid['provincia'] == 'Málaga']
data_Malaga = data_Malaga.set_index('fecha')
data_Malaga = data_Malaga.filter(['num_casos'])
data_Malaga = data_Malaga['2020-06-14':]
data_Malaga| num_casos | |
|---|---|
| fecha | |
| 2020-06-14 | 2 |
| 2020-06-15 | 1 |
| 2020-06-16 | 1 |
| 2020-06-17 | 2 |
| 2020-06-18 | 2 |
| ... | ... |
| 2022-03-25 | 565 |
| 2022-03-26 | 79 |
| 2022-03-27 | 65 |
| 2022-03-28 | 39 |
| 2022-03-29 | 0 |
654 rows × 1 columns
data_Malaga.describe()| num_casos | |
|---|---|
| count | 654.000000 |
| mean | 410.206422 |
| std | 489.452348 |
| min | 0.000000 |
| 25% | 122.250000 |
| 50% | 215.500000 |
| 75% | 475.250000 |
| max | 3080.000000 |
np_data_Malaga = data_Malaga.valuesscaler = MinMaxScaler(feature_range=(0, 1))
scaled_data_Malaga = scaler.fit_transform(np_data_Malaga)
print(f'Longitud del conjunto de datos disponible: {len(scaled_data_Malaga)}')Longitud del conjunto de datos disponible: 654# Since we are going to predict future values based on the X past elements,
# we need to create a list with those historic information for each element
historic_values = 90
scaled_data_Malaga_x = []
scaled_data_Malaga_y = []
for num_casos_i in range(historic_values, len(scaled_data_Malaga)):
scaled_data_Malaga_x.append(scaled_data_Malaga[(num_casos_i-historic_values):num_casos_i, 0])
scaled_data_Malaga_y.append(scaled_data_Malaga[num_casos_i, 0])
# Convert the x_train and y_train to numpy arrays
scaled_data_Malaga_x = np.array(scaled_data_Malaga_x)
scaled_data_Malaga_y = np.array(scaled_data_Malaga_y)# Train data looks like
scaled_data_Malaga_x[235]array([0.24448052, 0.21915584, 0.15649351, 0.12045455, 0.15357143,
0.14448052, 0.1474026 , 0.14675325, 0.11655844, 0.09448052,
0.06753247, 0.09545455, 0.11071429, 0.06883117, 0.06980519,
0.06688312, 0.04902597, 0.03474026, 0.05974026, 0.04935065,
0.04967532, 0.04480519, 0.04188312, 0.03993506, 0.02954545,
0.04188312, 0.04772727, 0.05162338, 0.04902597, 0.04318182,
0.02792208, 0.0288961 , 0.04253247, 0.04123377, 0.04025974,
0.03538961, 0.05097403, 0.03051948, 0.02662338, 0.04123377,
0.03149351, 0.04350649, 0.03733766, 0.03571429, 0.02954545,
0.0211039 , 0.04188312, 0.04545455, 0.03863636, 0.04772727,
0.04935065, 0.03571429, 0.0288961 , 0.05357143, 0.05324675,
0.05616883, 0.04253247, 0.04058442, 0.03474026, 0.04935065,
0.06623377, 0.07824675, 0.06623377, 0.05909091, 0.06233766,
0.05064935, 0.03116883, 0.07077922, 0.05649351, 0.07792208,
0.06168831, 0.0711039 , 0.04155844, 0.04577922, 0.06623377,
0.06720779, 0.05909091, 0.05519481, 0.05324675, 0.04383117,
0.03668831, 0.05941558, 0.06525974, 0.06006494, 0.05551948,
0.05876623, 0.04512987, 0.04545455, 0.06266234, 0.07727273])# Test data looks like
scaled_data_Malaga_y[235]0.0737012987012987# Since the first 90th values does not have historic, the dataset has been reduced in 90 values
print(f'Longitud datos de entrenamiento con historico: {len(scaled_data_Malaga_y)}')Longitud datos de entrenamiento con historico: 564# we split data in train and test
# as in previous analysis, we are going to predict a maximum of 91 days
x_train = scaled_data_Malaga_x[0:len(scaled_data_Malaga_x)-91]
y_train = scaled_data_Malaga_y[0:len(scaled_data_Malaga_y)-91]
print(f'Cantidad datos de entrenamiento: x={len(x_train)} - y={len(y_train)}')
x_test = scaled_data_Malaga_x[len(scaled_data_Malaga_x)-90:len(scaled_data_Malaga_x)]
y_test = scaled_data_Malaga_y[len(scaled_data_Malaga_y)-90:len(scaled_data_Malaga_y)]
print(f'Cantidad datos de test: x={len(x_test)} - y={len(y_test)}')Cantidad datos de entrenamiento: x=473 - y=473
Cantidad datos de test: x=90 - y=90# Reshape the data to feed de recurrent network
x_train = np.reshape(x_train, (x_train.shape[0], x_train.shape[1], 1))
print("Train data shape:")
print(x_train.shape)
print(y_train.shape)
x_test = np.reshape(x_test, (x_test.shape[0], x_test.shape[1], 1))
print("Test data shape:")
print(x_test.shape)
print(y_test.shape)Train data shape:
(473, 90, 1)
(473,)
Test data shape:
(90, 90, 1)
(90,)# Configure / setup the neural network model - LSTM
# Build the model
print('Build model...')
model = Sequential()
# Model with Neurons
# Inputshape = neurons -> Timestamps
neurons= x_train.shape[1]
model.add(LSTM(90,
activation = 'relu',
return_sequences = True,
input_shape = (x_train.shape[1], 1)))
model.add(LSTM(50,
activation = 'relu',
return_sequences = True))
model.add(LSTM(25,
activation = 'relu',
return_sequences = False))
model.add(Dense(5, activation = 'relu'))
model.add(Dense(1))Build model...model.compile(optimizer='adam', loss='mean_squared_error')
model.summary()Model: "sequential_3"_________________________________________________________________ Layer (type) Output Shape Param # ================================================================= lstm_9 (LSTM) (None, 90, 90) 33120 lstm_10 (LSTM) (None, 90, 50) 28200 lstm_11 (LSTM) (None, 25) 7600 dense_6 (Dense) (None, 5) 130 dense_7 (Dense) (None, 1) 6 =================================================================Total params: 69,056Trainable params: 69,056Non-trainable params: 0_________________________________________________________________# Training the model
# fit network
history = model.fit(x_train,
y_train,
batch_size = 1000,
epochs = 30,
validation_data = (x_test, y_test),
verbose = 0)plt.plot(history.history['loss'], label='train')
plt.plot(history.history['val_loss'], label='test')
plt.legend()
plt.show() 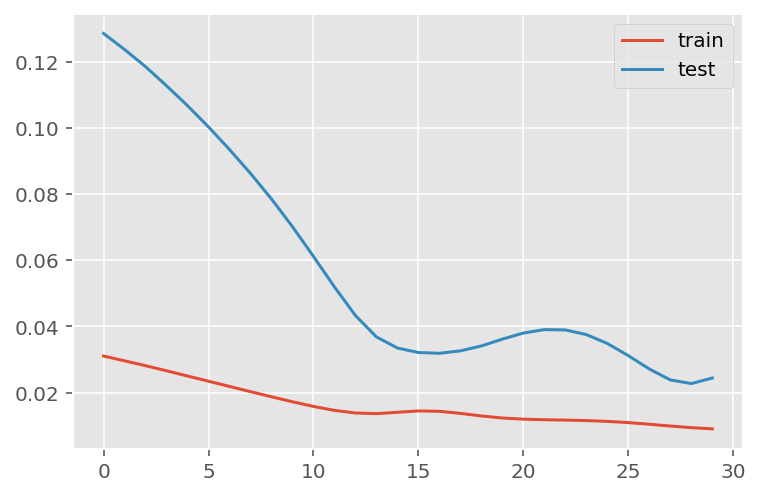
# Get the predicted values
predictions = model.predict(x_test)
predictions = scaler.inverse_transform(predictions)1/3 [=========>....................] - ETA: 0s3/3 [==============================] - ETA: 0s3/3 [==============================] - 0s 31ms/stepy_test = y_test.reshape(-1,1)
y_test = scaler.inverse_transform(y_test)# Calculate the mean absolute error (MAE)
mae = mean_absolute_error(y_test, predictions)
print('MAE: ' + str(round(mae, 1)))
# Calculate the root mean squarred error (RMSE)
rmse = np.sqrt(mean_squared_error(y_test,predictions))
print('RMSE: ' + str(round(rmse, 1)))
# Calculate the root mean squarred error (RMSE)
rmse = mean_squared_error(y_test,
predictions,
squared = False)
print('RMSE: ' + str(round(rmse, 1)))MAE: 353.5
RMSE: 481.1
RMSE: 481.1# Add the difference between the valid and predicted prices
train = data_Malaga[:(len(x_train)+92)]
valid = data_Malaga[(len(x_train)+91):]valid.insert(1, "Predictions", predictions, True)
valid.insert(1, "Difference", valid["Predictions"] - valid["num_casos"], True)# Zoom-in to a closer timeframe
# Date from which on the date is displayed
display_start_date = "2021-10-31"
valid = valid[valid.index > display_start_date]
train = train[train.index > display_start_date]# Visualize the data
matplotlib.style.use('ggplot')
fig, ax1 = plt.subplots(figsize=(22, 10), sharex=True)
# Data - Train
xt = train.index;
yt = train[["num_casos"]]
# Data - Test / validation
xv = valid.index;
yv = valid[["num_casos", "Predictions"]]
# Plot
plt.title("Malaga: Predictions vs Real infections", fontsize=20)
plt.ylabel("Nº Cases", fontsize=18)
plt.plot(yt, color="blue", linewidth=1.5)
plt.plot(yv["Predictions"], color="red", linewidth=1.5)
plt.plot(yv["num_casos"], color="green", linewidth=1.5)
plt.legend(["Train", "LSTM Predictions", "Test"],
loc="upper left", fontsize=18)
# Bar plot with the differences
x = valid.index
y = valid["Difference"]
plt.bar(x, y, width=0.2, color="grey")
plt.grid()
plt.show()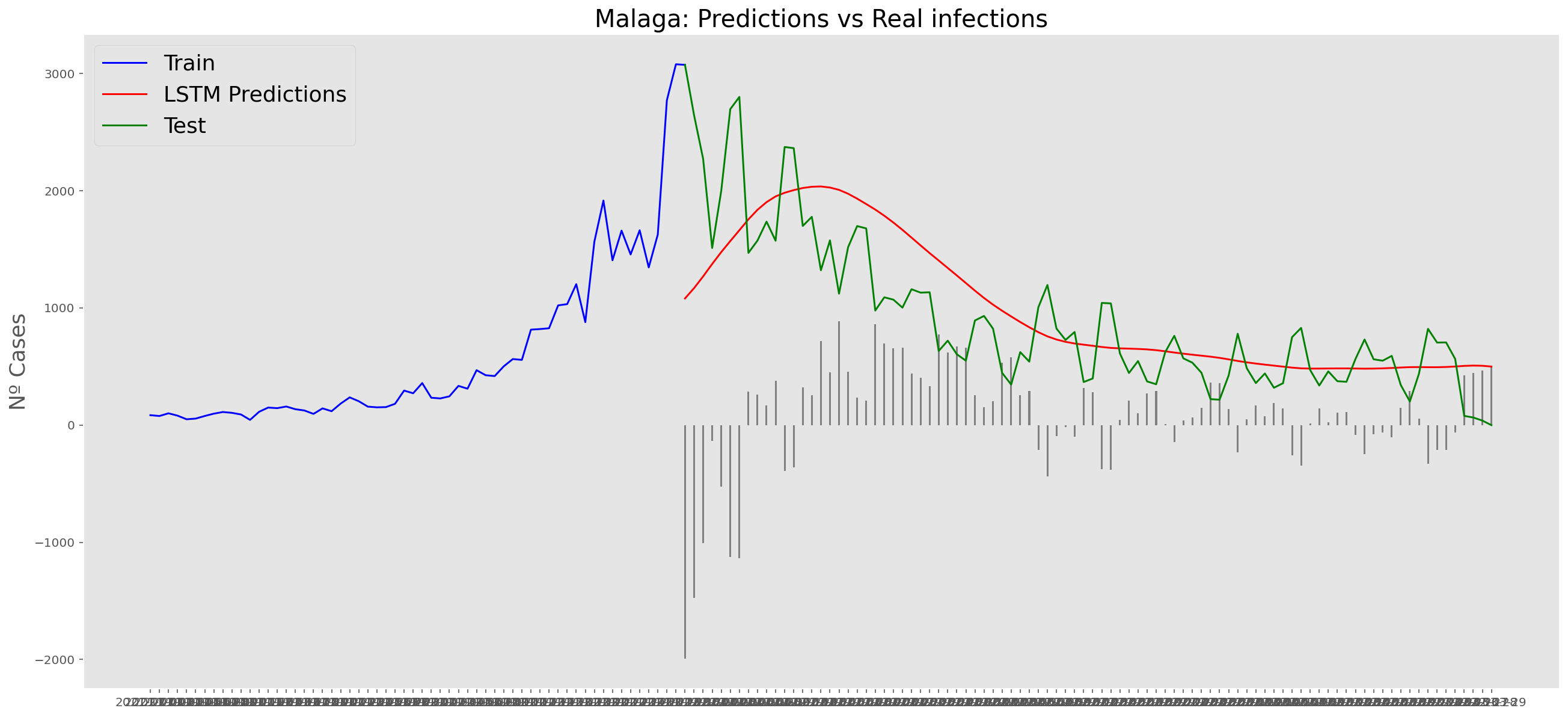
Sevilla
data_Sevilla = data_covid.loc[data_covid['provincia'] == 'Sevilla']
data_Sevilla = data_Sevilla.set_index('fecha')
data_Sevilla = data_Sevilla.filter(['num_casos'])
data_Sevilla = data_Sevilla['2020-06-14':]
data_Sevilla| num_casos | |
|---|---|
| fecha | |
| 2020-06-14 | 0 |
| 2020-06-15 | 2 |
| 2020-06-16 | 1 |
| 2020-06-17 | 0 |
| 2020-06-18 | 0 |
| ... | ... |
| 2022-03-25 | 365 |
| 2022-03-26 | 67 |
| 2022-03-27 | 24 |
| 2022-03-28 | 12 |
| 2022-03-29 | 0 |
654 rows × 1 columns
data_Sevilla.describe()| num_casos | |
|---|---|
| count | 654.000000 |
| mean | 432.978593 |
| std | 500.618517 |
| min | 0.000000 |
| 25% | 127.250000 |
| 50% | 282.000000 |
| 75% | 528.250000 |
| max | 3692.000000 |
np_data_Sevilla = data_Sevilla.valuesscaler = MinMaxScaler(feature_range=(0, 1))
scaled_data_Sevilla = scaler.fit_transform(np_data_Sevilla)
print(f'Longitud del conjunto de datos disponible: {len(scaled_data_Sevilla)}')Longitud del conjunto de datos disponible: 654# Since we are going to predict future values based on the X past elements,
# we need to create a list with those historic information for each element
historic_values = 90
scaled_data_Sevilla_x = []
scaled_data_Sevilla_y = []
for num_casos_i in range(historic_values, len(scaled_data_Sevilla)):
scaled_data_Sevilla_x.append(scaled_data_Sevilla[(num_casos_i-historic_values):num_casos_i, 0])
scaled_data_Sevilla_y.append(scaled_data_Sevilla[num_casos_i, 0])
# Convert the x_train and y_train to numpy arrays
scaled_data_Sevilla_x = np.array(scaled_data_Sevilla_x)
scaled_data_Sevilla_y = np.array(scaled_data_Sevilla_y)# Train data looks like
scaled_data_Sevilla_x[235]array([0.18634886, 0.19799567, 0.14626219, 0.0896533 , 0.13894908,
0.1695558 , 0.14572048, 0.12242687, 0.13434453, 0.06500542,
0.05390033, 0.08206934, 0.08667389, 0.09452871, 0.06798483,
0.05904659, 0.04252438, 0.0343987 , 0.06554713, 0.06473456,
0.05444204, 0.0503792 , 0.05796316, 0.04577465, 0.03819068,
0.04712893, 0.0528169 , 0.0663597 , 0.06013001, 0.05119177,
0.03764897, 0.031961 , 0.05119177, 0.05010834, 0.04739978,
0.05119177, 0.05390033, 0.03927411, 0.03737811, 0.06013001,
0.05633803, 0.05606717, 0.05850488, 0.06473456, 0.04658722,
0.04198267, 0.07150596, 0.07340195, 0.05823402, 0.08071506,
0.07773564, 0.05877573, 0.05471289, 0.07990249, 0.08829902,
0.0872156 , 0.09019502, 0.07069339, 0.08775731, 0.08396533,
0.14951246, 0.12432286, 0.12269772, 0.12378115, 0.13028169,
0.11348862, 0.07936078, 0.13299025, 0.12269772, 0.11213434,
0.12107259, 0.12432286, 0.09723727, 0.07042254, 0.09886241,
0.11782232, 0.10157096, 0.10346696, 0.12621885, 0.08342362,
0.07340195, 0.0847779 , 0.08559047, 0.11159263, 0.10292524,
0.08911159, 0.07502709, 0.04821235, 0.08315276, 0.09913326])# Test data looks like
scaled_data_Sevilla_y[235]0.10861321776814734# Since the first 90th values does not have historic, the dataset has been reduced in 90 values
print(f'Longitud datos de entrenamiento con historico: {len(scaled_data_Sevilla_y)}')Longitud datos de entrenamiento con historico: 564# we split data in train and test
# as in previous analysis, we are going to predict a maximum of 91 days
x_train = scaled_data_Sevilla_x[0:len(scaled_data_Sevilla_x)-91]
y_train = scaled_data_Sevilla_y[0:len(scaled_data_Sevilla_y)-91]
print(f'Cantidad datos de entrenamiento: x={len(x_train)} - y={len(y_train)}')
x_test = scaled_data_Sevilla_x[len(scaled_data_Sevilla_x)-90:len(scaled_data_Sevilla_x)]
y_test = scaled_data_Sevilla_y[len(scaled_data_Sevilla_y)-90:len(scaled_data_Sevilla_y)]
print(f'Cantidad datos de test: x={len(x_test)} - y={len(y_test)}')Cantidad datos de entrenamiento: x=473 - y=473
Cantidad datos de test: x=90 - y=90# Reshape the data to feed de recurrent network
x_train = np.reshape(x_train, (x_train.shape[0], x_train.shape[1], 1))
print("Train data shape:")
print(x_train.shape)
print(y_train.shape)
x_test = np.reshape(x_test, (x_test.shape[0], x_test.shape[1], 1))
print("Test data shape:")
print(x_test.shape)
print(y_test.shape)Train data shape:
(473, 90, 1)
(473,)
Test data shape:
(90, 90, 1)
(90,)# Configure / setup the neural network model - LSTM
# Build the model
print('Build model...')
model = Sequential()
# Model with Neurons
# Inputshape = neurons -> Timestamps
neurons= x_train.shape[1]
model.add(LSTM(90,
activation = 'relu',
return_sequences = True,
input_shape = (x_train.shape[1], 1)))
model.add(LSTM(50,
activation = 'relu',
return_sequences = True))
model.add(LSTM(25,
activation = 'relu',
return_sequences = False))
model.add(Dense(5, activation = 'relu'))
model.add(Dense(1))Build model...model.compile(optimizer='adam', loss='mean_squared_error')
model.summary()Model: "sequential_4"_________________________________________________________________ Layer (type) Output Shape Param # ================================================================= lstm_12 (LSTM) (None, 90, 90) 33120 lstm_13 (LSTM) (None, 90, 50) 28200 lstm_14 (LSTM) (None, 25) 7600 dense_8 (Dense) (None, 5) 130 dense_9 (Dense) (None, 1) 6 =================================================================Total params: 69,056Trainable params: 69,056Non-trainable params: 0_________________________________________________________________# Training the model
# fit network
history = model.fit(x_train,
y_train,
batch_size = 1000,
epochs = 30,
validation_data = (x_test, y_test),
verbose = 0)plt.plot(history.history['loss'], label='train')
plt.plot(history.history['val_loss'], label='test')
plt.legend()
plt.show() 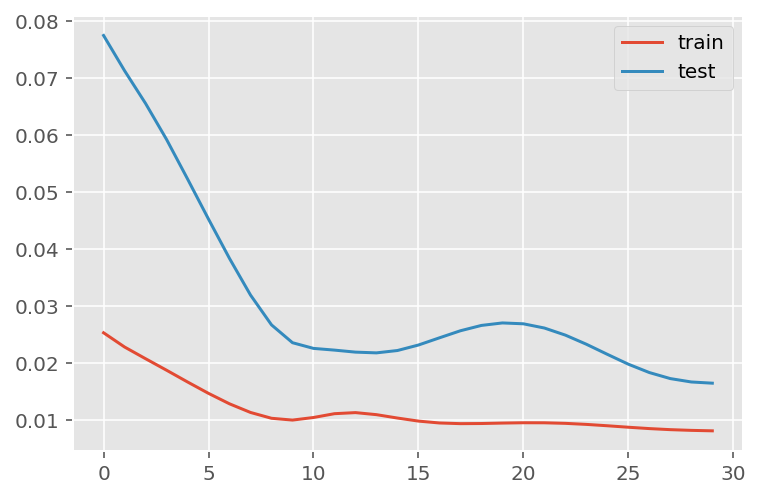
# Get the predicted values
predictions = model.predict(x_test)
predictions = scaler.inverse_transform(predictions)WARNING:tensorflow:5 out of the last 13 calls to <function Model.make_predict_function.<locals>.predict_function at 0x7fdc69aa8c10> triggered tf.function retracing. Tracing is expensive and the excessive number of tracings could be due to (1) creating @tf.function repeatedly in a loop, (2) passing tensors with different shapes, (3) passing Python objects instead of tensors. For (1), please define your @tf.function outside of the loop. For (2), @tf.function has reduce_retracing=True option that can avoid unnecessary retracing. For (3), please refer to https://www.tensorflow.org/guide/function#controlling_retracing and https://www.tensorflow.org/api_docs/python/tf/function for more details.1/3 [=========>....................] - ETA: 0s3/3 [==============================] - ETA: 0s3/3 [==============================] - 0s 32ms/stepy_test = y_test.reshape(-1,1)
y_test = scaler.inverse_transform(y_test)# Calculate the mean absolute error (MAE)
mae = mean_absolute_error(y_test, predictions)
print('MAE: ' + str(round(mae, 1)))
# Calculate the root mean squarred error (RMSE)
rmse = np.sqrt(mean_squared_error(y_test,predictions))
print('RMSE: ' + str(round(rmse, 1)))
# Calculate the root mean squarred error (RMSE)
rmse = mean_squared_error(y_test,
predictions,
squared = False)
print('RMSE: ' + str(round(rmse, 1)))MAE: 335.1
RMSE: 474.0
RMSE: 474.0# Add the difference between the valid and predicted prices
train = data_Sevilla[:(len(x_train)+92)]
valid = data_Sevilla[(len(x_train)+91):]valid.insert(1, "Predictions", predictions, True)
valid.insert(1, "Difference", valid["Predictions"] - valid["num_casos"], True)# Zoom-in to a closer timeframe
# Date from which on the date is displayed
display_start_date = "2021-10-31"
valid = valid[valid.index > display_start_date]
train = train[train.index > display_start_date]# Visualize the data
matplotlib.style.use('ggplot')
fig, ax1 = plt.subplots(figsize=(22, 10), sharex=True)
# Data - Train
xt = train.index;
yt = train[["num_casos"]]
# Data - Test / validation
xv = valid.index;
yv = valid[["num_casos", "Predictions"]]
# Plot
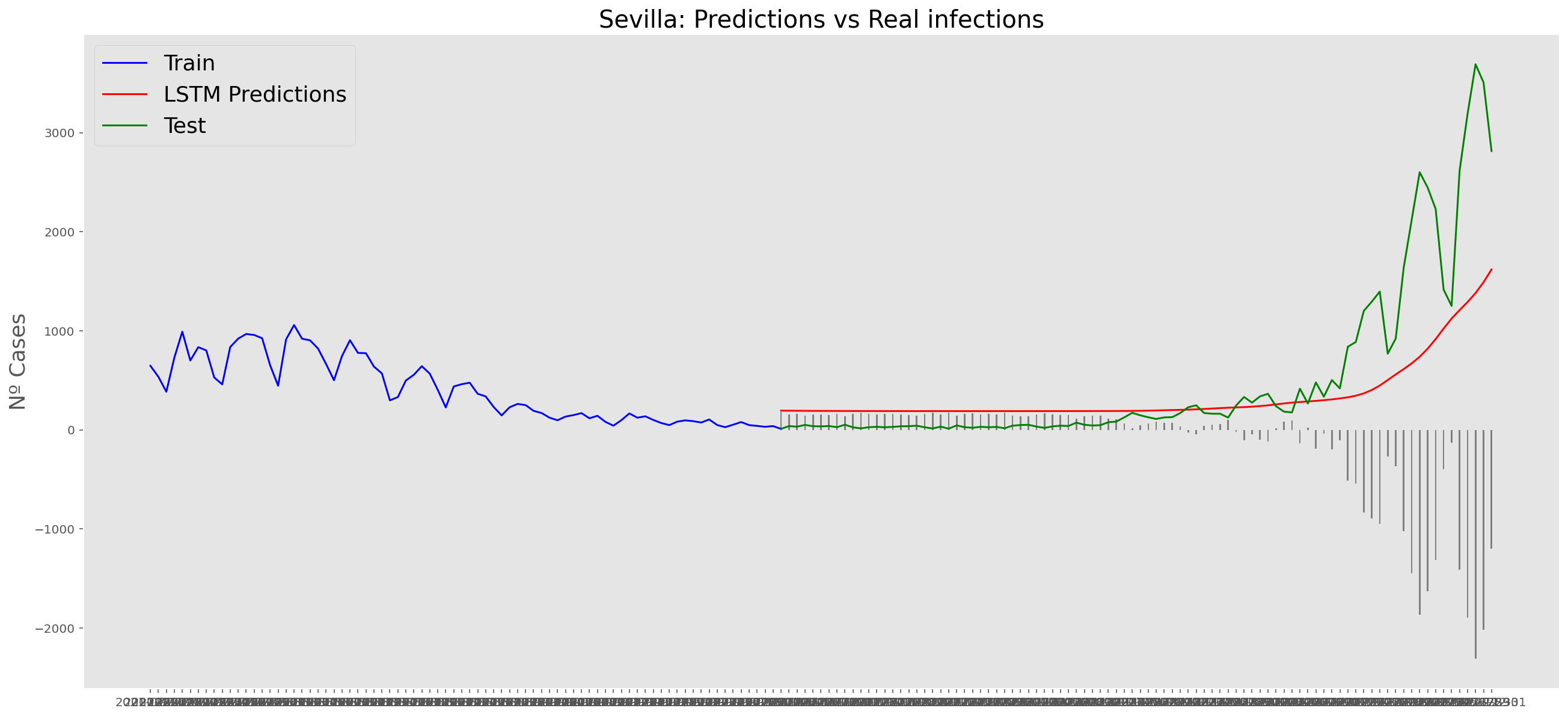
plt.title("Sevilla: Predictions vs Real infections", fontsize=20)
plt.ylabel("Nº Cases", fontsize=18)
plt.plot(yt, color="blue", linewidth=1.5)
plt.plot(yv["Predictions"], color="red", linewidth=1.5)
plt.plot(yv["num_casos"], color="green", linewidth=1.5)
plt.legend(["Train", "LSTM Predictions", "Test"],
loc="upper left", fontsize=18)
# Bar plot with the differences
x = valid.index
y = valid["Difference"]
plt.bar(x, y, width=0.2, color="grey")
plt.grid()
plt.show()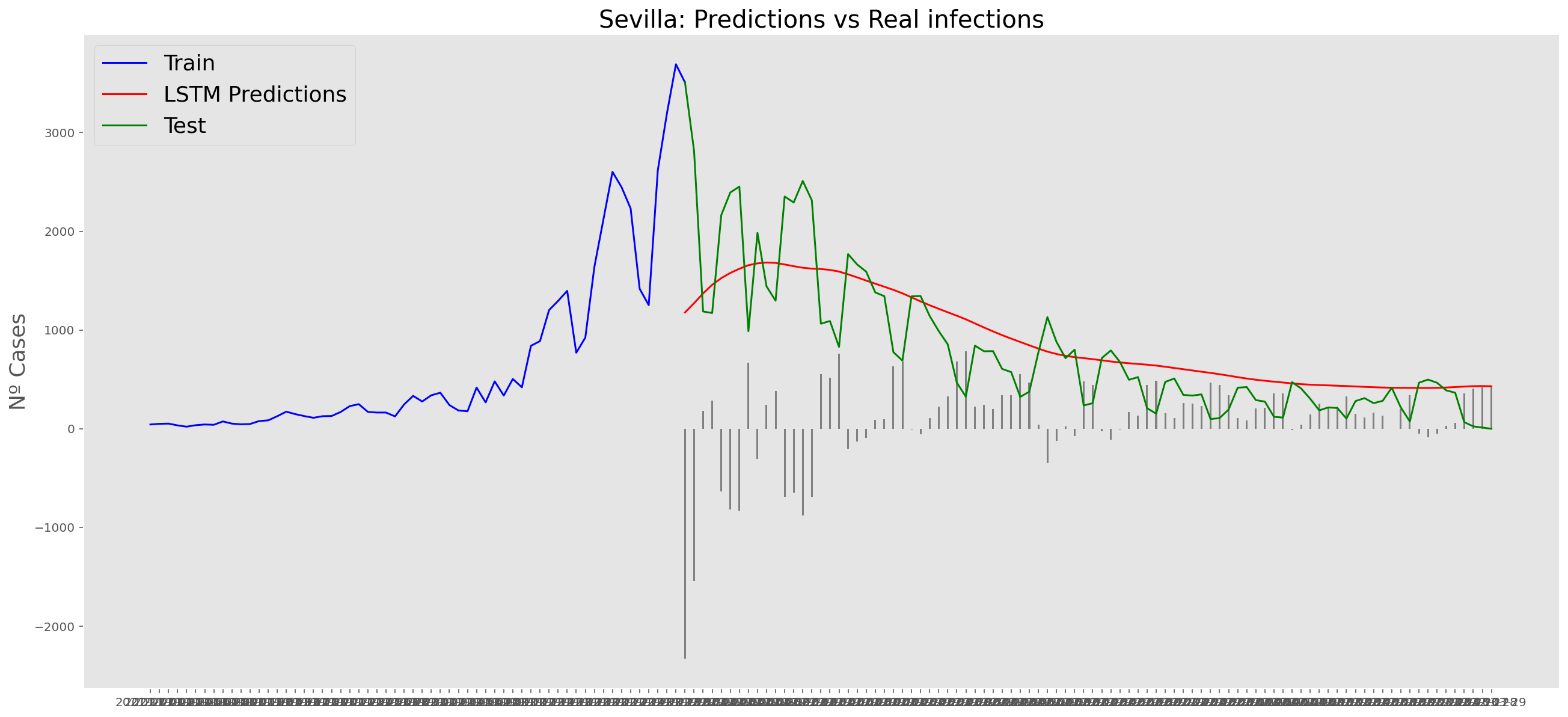
Just before the sixth wave
Asturias
data_asturias = data_covid.loc[data_covid['provincia'] == 'Asturias']
data_asturias = data_asturias.set_index('fecha')
data_asturias = data_asturias.filter(['num_casos'])
data_asturias = data_asturias['2020-06-14':'2021-12-31']
data_asturias| num_casos | |
|---|---|
| fecha | |
| 2020-06-14 | 0 |
| 2020-06-15 | 0 |
| 2020-06-16 | 1 |
| 2020-06-17 | 0 |
| 2020-06-18 | 0 |
| ... | ... |
| 2021-12-27 | 2363 |
| 2021-12-28 | 2150 |
| 2021-12-29 | 2159 |
| 2021-12-30 | 2020 |
| 2021-12-31 | 1949 |
566 rows × 1 columns
data_asturias.describe()| num_casos | |
|---|---|
| count | 566.000000 |
| mean | 180.913428 |
| std | 276.250633 |
| min | 0.000000 |
| 25% | 36.000000 |
| 50% | 94.500000 |
| 75% | 225.750000 |
| max | 2363.000000 |
np_data_asturias = data_asturias.valuesscaler = MinMaxScaler(feature_range=(0, 1))
scaled_data_asturias = scaler.fit_transform(np_data_asturias)
print(f'Longitud del conjunto de datos disponible: {len(scaled_data_asturias)}')Longitud del conjunto de datos disponible: 566# Since we are going to predict future values based on the X past elements,
# we need to create a list with those historic information for each element
historic_values = 90
scaled_data_asturias_x = []
scaled_data_asturias_y = []
for num_casos_i in range(historic_values, len(scaled_data_asturias)):
scaled_data_asturias_x.append(scaled_data_asturias[(num_casos_i-historic_values):num_casos_i, 0])
scaled_data_asturias_y.append(scaled_data_asturias[num_casos_i, 0])
# Convert the x_train and y_train to numpy arrays
scaled_data_asturias_x = np.array(scaled_data_asturias_x)
scaled_data_asturias_y = np.array(scaled_data_asturias_y)# Since the first 90 values does not have historic, the dataset has been reduced in 90 values
print(f'Longitud datos de entrenamiento con historico: {len(scaled_data_asturias_y)}')Longitud datos de entrenamiento con historico: 476# we split data in train and test
# as in previous analysis, we are going to predict a maximum of 21 days
x_train = scaled_data_asturias_x[0:len(scaled_data_asturias_x)-91]
y_train = scaled_data_asturias_y[0:len(scaled_data_asturias_y)-91]
print(f'Cantidad datos de entrenamiento: x={len(x_train)} - y={len(y_train)}')
x_test = scaled_data_asturias_x[len(scaled_data_asturias_x)-90:len(scaled_data_asturias_x)]
y_test = scaled_data_asturias_y[len(scaled_data_asturias_y)-90:len(scaled_data_asturias_y)]
print(f'Cantidad datos de test: x={len(x_test)} - y={len(y_test)}')Cantidad datos de entrenamiento: x=385 - y=385
Cantidad datos de test: x=90 - y=90# Reshape the data to feed de recurrent network
x_train = np.reshape(x_train, (x_train.shape[0], x_train.shape[1], 1))
print("Train data shape:")
print(x_train.shape)
print(y_train.shape)
x_test = np.reshape(x_test, (x_test.shape[0], x_test.shape[1], 1))
print("Test data shape:")
print(x_test.shape)
print(y_test.shape)Train data shape:
(385, 90, 1)
(385,)
Test data shape:
(90, 90, 1)
(90,)# # Configure / setup the neural network model - LSTM
# Build the model
print('Build model...')
model = Sequential()
# Model with Neurons
# Inputshape = neurons -> Timestamps
neurons= x_train.shape[1]
model.add(LSTM(90,
activation = 'relu',
return_sequences = True,
input_shape = (x_train.shape[1], 1)))
model.add(LSTM(50,
activation = 'relu',
return_sequences = True))
model.add(LSTM(25,
activation = 'relu',
return_sequences = False))
model.add(Dense(5, activation = 'relu'))
model.add(Dense(1))Build model...model.compile(optimizer='adam', loss='mean_squared_error')
model.summary()Model: "sequential_5"_________________________________________________________________ Layer (type) Output Shape Param # ================================================================= lstm_15 (LSTM) (None, 90, 90) 33120 lstm_16 (LSTM) (None, 90, 50) 28200 lstm_17 (LSTM) (None, 25) 7600 dense_10 (Dense) (None, 5) 130 dense_11 (Dense) (None, 1) 6 =================================================================Total params: 69,056Trainable params: 69,056Non-trainable params: 0_________________________________________________________________# Training the model
# # fit network
history = model.fit(x_train,
y_train,
batch_size=1000,
epochs=50,
validation_data = (x_test, y_test),
verbose = 0)plt.plot(history.history['loss'], label='train')
plt.plot(history.history['val_loss'], label='test')
plt.legend()
plt.show() 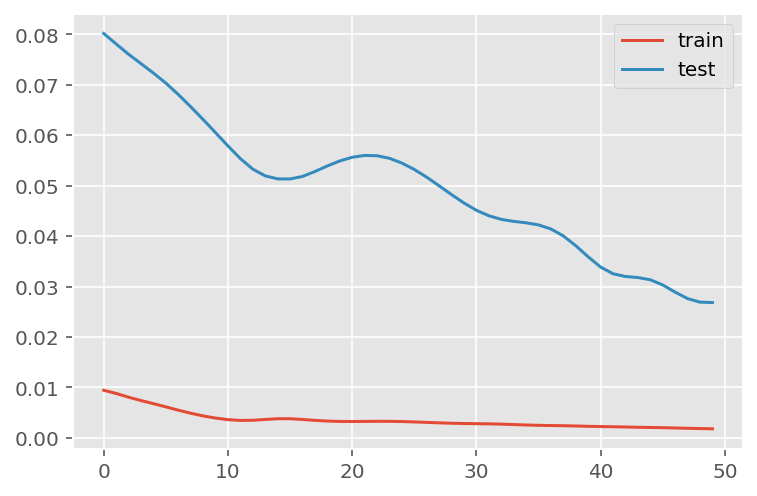
# Get the predicted values
predictions = model.predict(x_test)
predictions = scaler.inverse_transform(predictions)WARNING:tensorflow:5 out of the last 13 calls to <function Model.make_predict_function.<locals>.predict_function at 0x7fdc3a579c10> triggered tf.function retracing. Tracing is expensive and the excessive number of tracings could be due to (1) creating @tf.function repeatedly in a loop, (2) passing tensors with different shapes, (3) passing Python objects instead of tensors. For (1), please define your @tf.function outside of the loop. For (2), @tf.function has reduce_retracing=True option that can avoid unnecessary retracing. For (3), please refer to https://www.tensorflow.org/guide/function#controlling_retracing and https://www.tensorflow.org/api_docs/python/tf/function for more details.1/3 [=========>....................] - ETA: 0s3/3 [==============================] - ETA: 0s3/3 [==============================] - 0s 31ms/stepy_test = y_test.reshape(-1,1)
y_test = scaler.inverse_transform(y_test)# Calculate the mean absolute error (MAE)
mae = mean_absolute_error(y_test, predictions)
print('MAE: ' + str(round(mae, 1)))
# Calculate the root mean squarred error (RMSE)
rmse = np.sqrt(mean_squared_error(y_test,predictions))
print('RMSE: ' + str(round(rmse, 1)))
# Calculate the root mean squarred error (RMSE)
rmse = mean_squared_error(y_test,
predictions,
squared = False)
print('RMSE: ' + str(round(rmse, 1)))MAE: 208.7
RMSE: 387.0
RMSE: 387.0# Add the difference between the valid and predicted prices
train = data_asturias[:(len(x_train)+92)]
valid = data_asturias[(len(x_train)+91):]valid.insert(1, "Predictions", predictions, True)
valid.insert(1, "Difference", valid["Predictions"] - valid["num_casos"], True)# Zoom-in to a closer timeframe
# Date from which on the date is displayed
display_start_date = "2021-07-15"
valid = valid[valid.index > display_start_date]
train = train[train.index > display_start_date]# Visualize the data
matplotlib.style.use('ggplot')
fig, ax1 = plt.subplots(figsize=(22, 10), sharex=True)
# Data - Train
xt = train.index;
yt = train[["num_casos"]]
# Data - Test / validation
xv = valid.index;
yv = valid[["num_casos", "Predictions"]]
# Plot
plt.title("Asturias: Predictions vs Real infections", fontsize=20)
plt.ylabel("Nº Cases", fontsize=18)
plt.plot(yt, color="blue", linewidth=1.5)
plt.plot(yv["Predictions"], color="red", linewidth=1.5)
plt.plot(yv["num_casos"], color="green", linewidth=1.5)
plt.legend(["Train", "LSTM Predictions", "Test"],
loc="upper left", fontsize=18)
# Bar plot with the differences
x = valid.index
y = valid["Difference"]
plt.bar(x, y, width=0.2, color="grey")
plt.grid()
plt.show()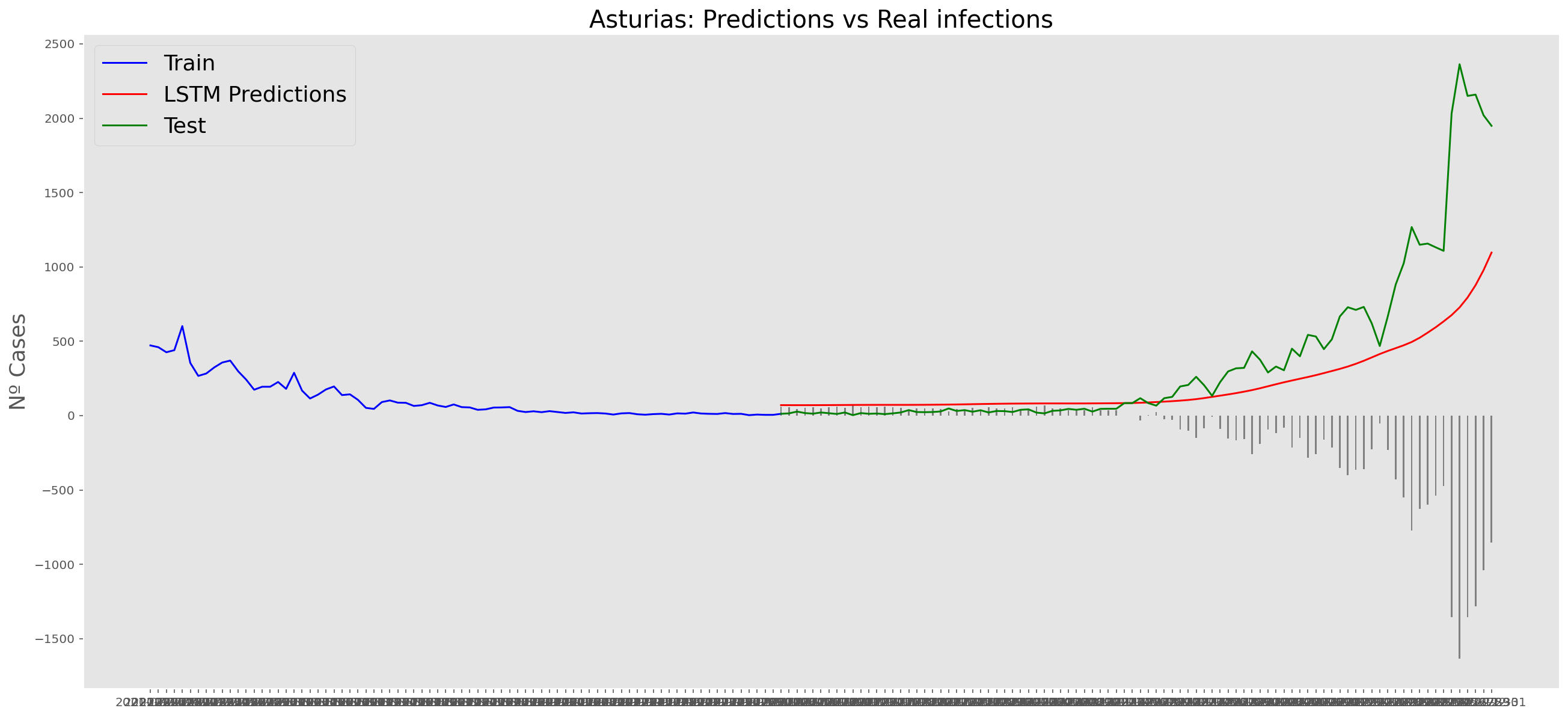
Barcelona
data_Barcelona = data_covid.loc[data_covid['provincia'] == 'Barcelona']
data_Barcelona = data_Barcelona.set_index('fecha')
data_Barcelona = data_Barcelona.filter(['num_casos'])
data_Barcelona = data_Barcelona['2020-06-14':'2021-12-31']
data_Barcelona| num_casos | |
|---|---|
| fecha | |
| 2020-06-14 | 33 |
| 2020-06-15 | 62 |
| 2020-06-16 | 66 |
| 2020-06-17 | 70 |
| 2020-06-18 | 68 |
| ... | ... |
| 2021-12-27 | 19383 |
| 2021-12-28 | 20192 |
| 2021-12-29 | 19361 |
| 2021-12-30 | 17639 |
| 2021-12-31 | 16651 |
566 rows × 1 columns
data_Barcelona.describe()| num_casos | |
|---|---|
| count | 566.000000 |
| mean | 1575.480565 |
| std | 2281.290383 |
| min | 33.000000 |
| 25% | 544.750000 |
| 50% | 944.500000 |
| 75% | 1609.500000 |
| max | 20192.000000 |
np_data_Barcelona = data_Barcelona.valuesscaler = MinMaxScaler(feature_range=(0, 1))
scaled_data_Barcelona = scaler.fit_transform(np_data_Barcelona)
print(f'Longitud del conjunto de datos disponible: {len(scaled_data_Barcelona)}')Longitud del conjunto de datos disponible: 566# Since we are going to predict future values based on the 90 past elements,
# we need to create a list with those historic information for each element
historic_values = 90
scaled_data_Barcelona_x = []
scaled_data_Barcelona_y = []
for num_casos_i in range(historic_values, len(scaled_data_Barcelona)):
scaled_data_Barcelona_x.append(scaled_data_Barcelona[(num_casos_i-historic_values):num_casos_i, 0])
scaled_data_Barcelona_y.append(scaled_data_Barcelona[num_casos_i, 0])
# Convert the x_train and y_train to numpy arrays
scaled_data_Barcelona_x = np.array(scaled_data_Barcelona_x)
scaled_data_Barcelona_y = np.array(scaled_data_Barcelona_y)# Train data looks like
scaled_data_Barcelona_x[235]array([0.08507366, 0.07971626, 0.04826628, 0.04151992, 0.07822809,
0.07048961, 0.06289995, 0.05908031, 0.06354482, 0.04345454,
0.03884121, 0.06855499, 0.06309837, 0.06012203, 0.06116375,
0.05208592, 0.0405278 , 0.03422789, 0.06364403, 0.06056848,
0.05337566, 0.045786 , 0.04687733, 0.03323578, 0.02852324,
0.05833623, 0.05064735, 0.04777023, 0.03864279, 0.04330572,
0.03065628, 0.02668783, 0.04707575, 0.04355375, 0.04449625,
0.04251203, 0.04305769, 0.03199563, 0.02842403, 0.05317724,
0.04955603, 0.04266085, 0.04722456, 0.04861352, 0.0360633 ,
0.03090431, 0.06240389, 0.06106454, 0.05441738, 0.05074656,
0.05957637, 0.04181755, 0.03536882, 0.06384245, 0.05734411,
0.05084578, 0.0563024 , 0.03655935, 0.04191676, 0.04196637,
0.04712535, 0.06438811, 0.06275113, 0.05883228, 0.06170941,
0.0475222 , 0.03482316, 0.0651322 , 0.05873307, 0.06696761,
0.05268118, 0.05679845, 0.04008135, 0.03571606, 0.06642195,
0.077484 , 0.05863386, 0.06652116, 0.0546158 , 0.04608364,
0.03933727, 0.07872414, 0.07088645, 0.06389206, 0.06156059,
0.05878268, 0.03462473, 0.02753113, 0.06384245, 0.04682772])# Test data looks like
scaled_data_Barcelona_y[235]0.05114340989136366# Since the first 90th values does not have historic, the dataset has been reduced in 90 values
print(f'Longitud datos de entrenamiento con historico: {len(scaled_data_Barcelona_y)}')Longitud datos de entrenamiento con historico: 476# we split data in train and test
# as in previous analysis, we are going to predict a maximum of 90 days
x_train = scaled_data_Barcelona_x[0:len(scaled_data_Barcelona_x)-91]
y_train = scaled_data_Barcelona_y[0:len(scaled_data_Barcelona_y)-91]
print(f'Cantidad datos de entrenamiento: x={len(x_train)} - y={len(y_train)}')
x_test = scaled_data_Barcelona_x[len(scaled_data_Barcelona_x)-90:len(scaled_data_Barcelona_x)]
y_test = scaled_data_Barcelona_y[len(scaled_data_Barcelona_y)-90:len(scaled_data_Barcelona_y)]
print(f'Cantidad datos de test: x={len(x_test)} - y={len(y_test)}')Cantidad datos de entrenamiento: x=385 - y=385
Cantidad datos de test: x=90 - y=90# Reshape the data to feed de recurrent network
x_train = np.reshape(x_train, (x_train.shape[0], x_train.shape[1], 1))
print("Train data shape:")
print(x_train.shape)
print(y_train.shape)
x_test = np.reshape(x_test, (x_test.shape[0], x_test.shape[1], 1))
print("Test data shape:")
print(x_test.shape)
print(y_test.shape)Train data shape:
(385, 90, 1)
(385,)
Test data shape:
(90, 90, 1)
(90,)# Configure / setup the neural network model - LSTM
# Build the model
print('Build model...')
model = Sequential()
# Model with Neurons
# Inputshape = neurons -> Timestamps
neurons= x_train.shape[1]
model.add(LSTM(90,
activation = 'relu',
return_sequences = True,
input_shape = (x_train.shape[1], 1)))
model.add(LSTM(50,
activation = 'relu',
return_sequences = True))
model.add(LSTM(25,
activation = 'relu',
return_sequences = False))
model.add(Dense(5, activation = 'relu'))
model.add(Dense(1))Build model...model.compile(optimizer='adam', loss='mean_squared_error')
model.summary()Model: "sequential_6"_________________________________________________________________ Layer (type) Output Shape Param # ================================================================= lstm_18 (LSTM) (None, 90, 90) 33120 lstm_19 (LSTM) (None, 90, 50) 28200 lstm_20 (LSTM) (None, 25) 7600 dense_12 (Dense) (None, 5) 130 dense_13 (Dense) (None, 1) 6 =================================================================Total params: 69,056Trainable params: 69,056Non-trainable params: 0_________________________________________________________________# Training the model
# fit network
history = model.fit(x_train,
y_train,
batch_size = 1000,
epochs = 50,
validation_data = (x_test, y_test),
verbose = 0)plt.plot(history.history['loss'], label='train')
plt.plot(history.history['val_loss'], label='test')
plt.legend()
plt.show() 
# Get the predicted values
predictions = model.predict(x_test)
predictions = scaler.inverse_transform(predictions)1/3 [=========>....................] - ETA: 0s3/3 [==============================] - ETA: 0s3/3 [==============================] - 0s 31ms/stepy_test = y_test.reshape(-1,1)
y_test = scaler.inverse_transform(y_test)# Calculate the mean absolute error (MAE)
mae = mean_absolute_error(y_test, predictions)
print('MAE: ' + str(round(mae, 1)))
# Calculate the root mean squarred error (RMSE)
rmse = np.sqrt(mean_squared_error(y_test,predictions))
print('RMSE: ' + str(round(rmse, 1)))
# Calculate the root mean squarred error (RMSE)
rmse = mean_squared_error(y_test,
predictions,
squared = False)
print('RMSE: ' + str(round(rmse, 1)))MAE: 1674.3
RMSE: 3525.8
RMSE: 3525.8# Add the difference between the valid and predicted prices
train = data_Barcelona[:(len(x_train)+92)]
valid = data_Barcelona[(len(x_train)+91):]valid.insert(1, "Predictions", predictions, True)
valid.insert(1, "Difference", valid["Predictions"] - valid["num_casos"], True)# Zoom-in to a closer timeframe
# Date from which on the date is displayed
display_start_date = "2021-07-15"
valid = valid[valid.index > display_start_date]
train = train[train.index > display_start_date]# Visualize the data
matplotlib.style.use('ggplot')
fig, ax1 = plt.subplots(figsize=(22, 10), sharex=True)
# Data - Train
xt = train.index;
yt = train[["num_casos"]]
# Data - Test / validation
xv = valid.index;
yv = valid[["num_casos", "Predictions"]]
# Plot
plt.title("Barcelona: Predictions vs Real infections", fontsize=20)
plt.ylabel("Nº Cases", fontsize=18)
plt.plot(yt, color="blue", linewidth=1.5)
plt.plot(yv["Predictions"], color="red", linewidth=1.5)
plt.plot(yv["num_casos"], color="green", linewidth=1.5)
plt.legend(["Train", "LSTM Predictions", "Test"],
loc="upper left", fontsize=18)
# Bar plot with the differences
x = valid.index
y = valid["Difference"]
plt.bar(x, y, width=0.2, color="grey")
plt.grid()
plt.show()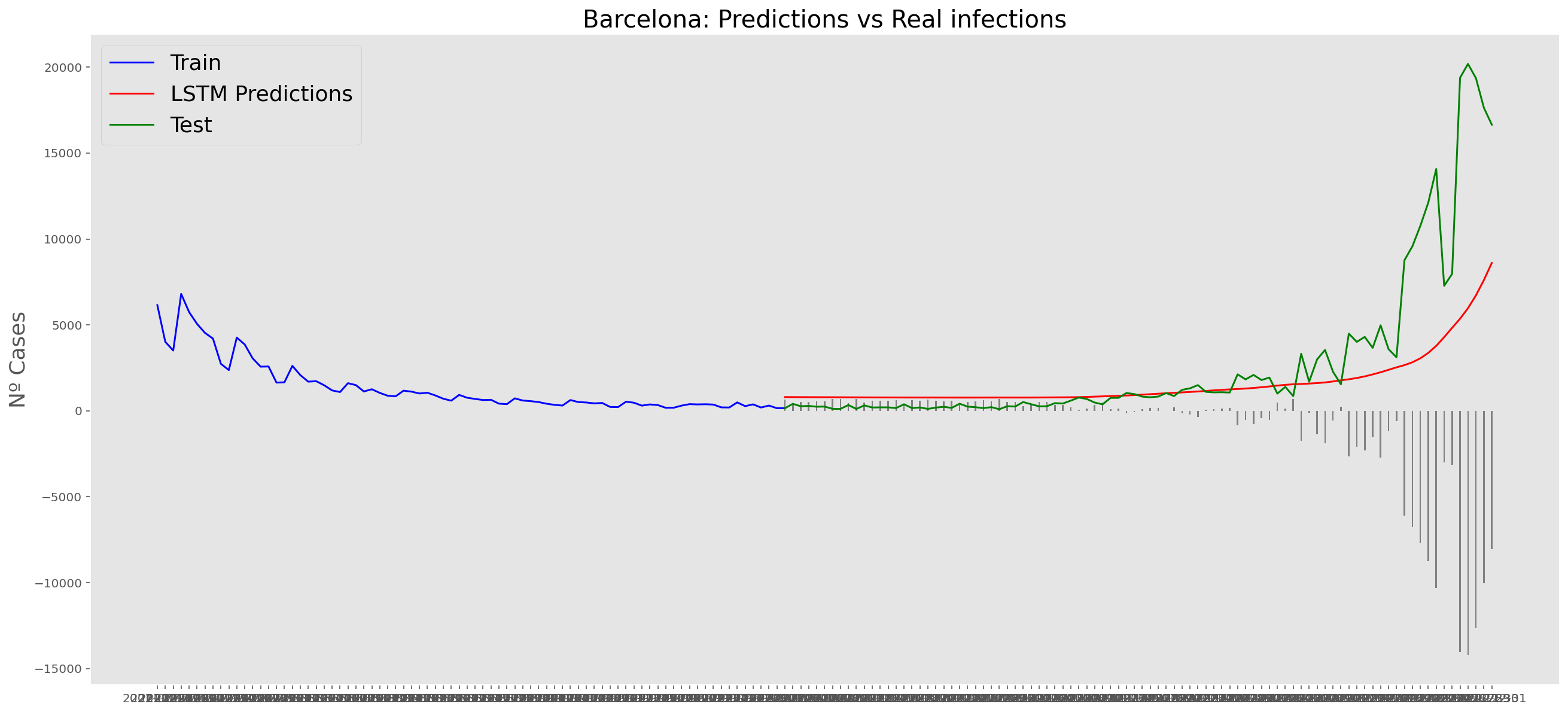
Madrid
data_Madrid = data_covid.loc[data_covid['provincia'] == 'Madrid']
data_Madrid = data_Madrid.set_index('fecha')
data_Madrid = data_Madrid.filter(['num_casos'])
data_Madrid = data_Madrid['2020-06-14':'2021-12-31']
data_Madrid| num_casos | |
|---|---|
| fecha | |
| 2020-06-14 | 81 |
| 2020-06-15 | 153 |
| 2020-06-16 | 91 |
| 2020-06-17 | 93 |
| 2020-06-18 | 85 |
| ... | ... |
| 2021-12-27 | 22958 |
| 2021-12-28 | 23811 |
| 2021-12-29 | 21914 |
| 2021-12-30 | 20666 |
| 2021-12-31 | 7556 |
566 rows × 1 columns
data_Madrid.describe()| num_casos | |
|---|---|
| count | 566.000000 |
| mean | 1994.136042 |
| std | 2795.419848 |
| min | 28.000000 |
| 25% | 550.250000 |
| 50% | 1312.500000 |
| 75% | 2282.500000 |
| max | 23811.000000 |
np_data_Madrid = data_Madrid.valuesscaler = MinMaxScaler(feature_range=(0, 1))
scaled_data_Madrid = scaler.fit_transform(np_data_Madrid)
print(f'Longitud del conjunto de datos disponible: {len(scaled_data_Madrid)}')Longitud del conjunto de datos disponible: 566# Since we are going to predict future values based on the X past elements,
# we need to create a list with those historic information for each element
historic_values = 90
scaled_data_Madrid_x = []
scaled_data_Madrid_y = []
for num_casos_i in range(historic_values, len(scaled_data_Madrid)):
scaled_data_Madrid_x.append(scaled_data_Madrid[(num_casos_i-historic_values):num_casos_i, 0])
scaled_data_Madrid_y.append(scaled_data_Madrid[num_casos_i, 0])
# Convert the x_train and y_train to numpy arrays
scaled_data_Madrid_x = np.array(scaled_data_Madrid_x)
scaled_data_Madrid_y = np.array(scaled_data_Madrid_y)# Train data looks like
scaled_data_Madrid_x[235]array([0.11428331, 0.14741622, 0.08796199, 0.06395324, 0.11087752,
0.11272758, 0.08863474, 0.06862044, 0.09216667, 0.06046336,
0.04620948, 0.08363117, 0.07660934, 0.06508851, 0.05621663,
0.06428962, 0.04747088, 0.04288778, 0.06286003, 0.05844511,
0.0505403 , 0.0445276 , 0.05243241, 0.04061725, 0.03943994,
0.05398814, 0.05138124, 0.04734474, 0.04171047, 0.05159147,
0.03830467, 0.03771602, 0.05474499, 0.05365177, 0.04726065,
0.04360257, 0.05201194, 0.04036497, 0.03906151, 0.05360972,
0.05096077, 0.05226422, 0.05625867, 0.04229912, 0.04974141,
0.04700837, 0.07206828, 0.07745028, 0.06176681, 0.05815078,
0.07400244, 0.05239036, 0.05247446, 0.07026027, 0.07009208,
0.07698776, 0.05381996, 0.06079973, 0.07063869, 0.07370811,
0.09796914, 0.09767481, 0.08493462, 0.08094017, 0.08867679,
0.07139553, 0.05890762, 0.09666569, 0.09679183, 0.08846655,
0.08459824, 0.09405878, 0.07101711, 0.0631964 , 0.092461 ,
0.09771686, 0.08350502, 0.07568431, 0.08821427, 0.06386915,
0.05398814, 0.08152882, 0.0865324 , 0.07240466, 0.06172476,
0.07400244, 0.04566287, 0.04406509, 0.04633562, 0.06613968])# Test data looks like
scaled_data_Madrid_y[235]0.0639532439137199# Since the first 90th values does not have historic, the dataset has been reduced in 90 values
print(f'Longitud datos de entrenamiento con historico: {len(scaled_data_Madrid_y)}')Longitud datos de entrenamiento con historico: 476# we split data in train and test
# as in previous analysis, we are going to predict a maximum of 91 days
x_train = scaled_data_Madrid_x[0:len(scaled_data_Madrid_x)-91]
y_train = scaled_data_Madrid_y[0:len(scaled_data_Madrid_y)-91]
print(f'Cantidad datos de entrenamiento: x={len(x_train)} - y={len(y_train)}')
x_test = scaled_data_Madrid_x[len(scaled_data_Madrid_x)-90:len(scaled_data_Madrid_x)]
y_test = scaled_data_Madrid_y[len(scaled_data_Madrid_y)-90:len(scaled_data_Madrid_y)]
print(f'Cantidad datos de test: x={len(x_test)} - y={len(y_test)}')Cantidad datos de entrenamiento: x=385 - y=385
Cantidad datos de test: x=90 - y=90# Reshape the data to feed de recurrent network
x_train = np.reshape(x_train, (x_train.shape[0], x_train.shape[1], 1))
print("Train data shape:")
print(x_train.shape)
print(y_train.shape)
x_test = np.reshape(x_test, (x_test.shape[0], x_test.shape[1], 1))
print("Test data shape:")
print(x_test.shape)
print(y_test.shape)Train data shape:
(385, 90, 1)
(385,)
Test data shape:
(90, 90, 1)
(90,)# Configure / setup the neural network model - LSTM
# Build the model
print('Build model...')
model = Sequential()
# Model with Neurons
# Inputshape = neurons -> Timestamps
neurons= x_train.shape[1]
model.add(LSTM(90,
activation = 'relu',
return_sequences = True,
input_shape = (x_train.shape[1], 1)))
model.add(LSTM(50,
activation = 'relu',
return_sequences = True))
model.add(LSTM(25,
activation = 'relu',
return_sequences = False))
model.add(Dense(5, activation = 'relu'))
model.add(Dense(1))Build model...model.compile(optimizer='adam', loss='mean_squared_error')
model.summary()Model: "sequential_7"_________________________________________________________________ Layer (type) Output Shape Param # ================================================================= lstm_21 (LSTM) (None, 90, 90) 33120 lstm_22 (LSTM) (None, 90, 50) 28200 lstm_23 (LSTM) (None, 25) 7600 dense_14 (Dense) (None, 5) 130 dense_15 (Dense) (None, 1) 6 =================================================================Total params: 69,056Trainable params: 69,056Non-trainable params: 0_________________________________________________________________# Training the model
# fit network
history = model.fit(x_train,
y_train,
batch_size = 1000,
epochs = 50,
validation_data = (x_test, y_test),
verbose = 0)plt.plot(history.history['loss'], label='train')
plt.plot(history.history['val_loss'], label='test')
plt.legend()
plt.show() 
# Get the predicted values
predictions = model.predict(x_test)
predictions = scaler.inverse_transform(predictions)1/3 [=========>....................] - ETA: 0s3/3 [==============================] - ETA: 0s3/3 [==============================] - 0s 31ms/stepy_test = y_test.reshape(-1,1)
y_test = scaler.inverse_transform(y_test)# Calculate the mean absolute error (MAE)
mae = mean_absolute_error(y_test, predictions)
print('MAE: ' + str(round(mae, 1)))
# Calculate the root mean squarred error (RMSE)
rmse = np.sqrt(mean_squared_error(y_test,predictions))
print('RMSE: ' + str(round(rmse, 1)))
# Calculate the root mean squarred error (RMSE)
rmse = mean_squared_error(y_test,
predictions,
squared = False)
print('RMSE: ' + str(round(rmse, 1)))MAE: 2521.6
RMSE: 5345.2
RMSE: 5345.2# Add the difference between the valid and predicted prices
train = data_Madrid[:(len(x_train)+92)]
valid = data_Madrid[(len(x_train)+91):]valid.insert(1, "Predictions", predictions, True)
valid.insert(1, "Difference", valid["Predictions"] - valid["num_casos"], True)# Zoom-in to a closer timeframe
# Date from which on the date is displayed
display_start_date = "2021-07-15"
valid = valid[valid.index > display_start_date]
train = train[train.index > display_start_date]# Visualize the data
matplotlib.style.use('ggplot')
fig, ax1 = plt.subplots(figsize=(22, 10), sharex=True)
# Data - Train
xt = train.index;
yt = train[["num_casos"]]
# Data - Test / validation
xv = valid.index;
yv = valid[["num_casos", "Predictions"]]
# Plot
plt.title("Madrid: Predictions vs Real infections", fontsize=20)
plt.ylabel("Nº Cases", fontsize=18)
plt.plot(yt, color="blue", linewidth=1.5)
plt.plot(yv["Predictions"], color="red", linewidth=1.5)
plt.plot(yv["num_casos"], color="green", linewidth=1.5)
plt.legend(["Train", "LSTM Predictions", "Test"],
loc="upper left", fontsize=18)
# Bar plot with the differences
x = valid.index
y = valid["Difference"]
plt.bar(x, y, width=0.2, color="grey")
plt.grid()
plt.show()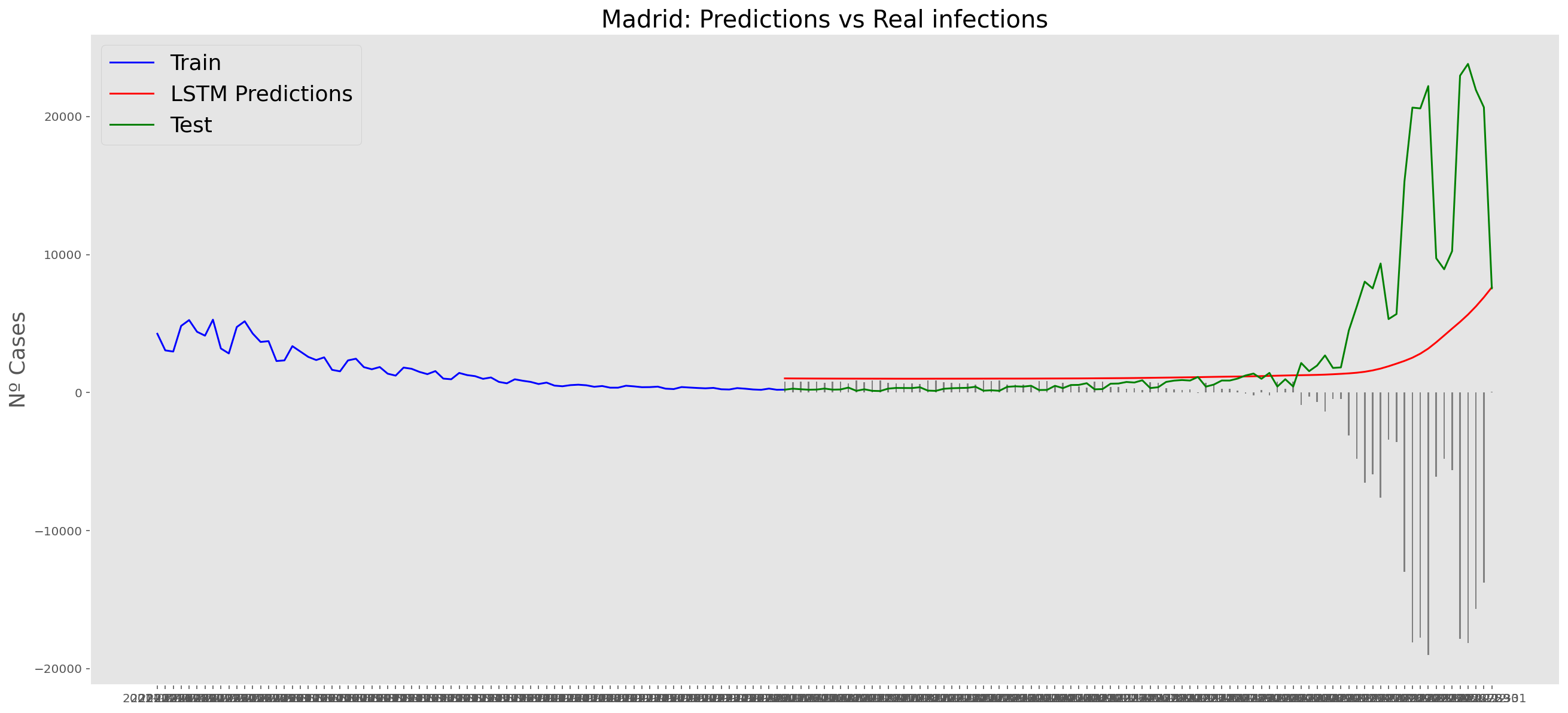
Malaga
data_Malaga = data_covid.loc[data_covid['provincia'] == 'Málaga']
data_Malaga = data_Malaga.set_index('fecha')
data_Malaga = data_Malaga.filter(['num_casos'])
data_Malaga = data_Malaga['2020-06-14':'2021-12-31']
data_Malaga| num_casos | |
|---|---|
| fecha | |
| 2020-06-14 | 2 |
| 2020-06-15 | 1 |
| 2020-06-16 | 1 |
| 2020-06-17 | 2 |
| 2020-06-18 | 2 |
| ... | ... |
| 2021-12-27 | 1627 |
| 2021-12-28 | 2772 |
| 2021-12-29 | 3080 |
| 2021-12-30 | 3075 |
| 2021-12-31 | 2646 |
566 rows × 1 columns
data_Malaga.describe()| num_casos | |
|---|---|
| count | 566.000000 |
| mean | 339.227915 |
| std | 427.261346 |
| min | 1.000000 |
| 25% | 110.000000 |
| 50% | 193.500000 |
| 75% | 344.750000 |
| max | 3080.000000 |
np_data_Malaga = data_Malaga.valuesscaler = MinMaxScaler(feature_range=(0, 1))
scaled_data_Malaga = scaler.fit_transform(np_data_Malaga)
print(f'Longitud del conjunto de datos disponible: {len(scaled_data_Malaga)}')Longitud del conjunto de datos disponible: 566# Since we are going to predict future values based on the X past elements,
# we need to create a list with those historic information for each element
historic_values = 90
scaled_data_Malaga_x = []
scaled_data_Malaga_y = []
for num_casos_i in range(historic_values, len(scaled_data_Malaga)):
scaled_data_Malaga_x.append(scaled_data_Malaga[(num_casos_i-historic_values):num_casos_i, 0])
scaled_data_Malaga_y.append(scaled_data_Malaga[num_casos_i, 0])
# Convert the x_train and y_train to numpy arrays
scaled_data_Malaga_x = np.array(scaled_data_Malaga_x)
scaled_data_Malaga_y = np.array(scaled_data_Malaga_y)# Train data looks like
scaled_data_Malaga_x[235]array([0.24423514, 0.21890224, 0.15621955, 0.12016889, 0.15329652,
0.14420266, 0.14712569, 0.14647613, 0.11627152, 0.09418642,
0.06722962, 0.09516077, 0.11042546, 0.06852874, 0.06950309,
0.06658006, 0.04871712, 0.03442676, 0.05943488, 0.0490419 ,
0.04936668, 0.04449497, 0.04157194, 0.03962325, 0.02923027,
0.04157194, 0.04741799, 0.05131536, 0.04871712, 0.04287106,
0.02760637, 0.02858071, 0.0422215 , 0.04092238, 0.03994804,
0.03507632, 0.0506658 , 0.03020461, 0.02630724, 0.04092238,
0.03117895, 0.04319584, 0.03702501, 0.0354011 , 0.02923027,
0.02078597, 0.04157194, 0.04514453, 0.03832413, 0.04741799,
0.0490419 , 0.0354011 , 0.02858071, 0.05326405, 0.05293927,
0.05586229, 0.0422215 , 0.04027282, 0.03442676, 0.0490419 ,
0.0659305 , 0.07794739, 0.0659305 , 0.05878532, 0.06203313,
0.05034102, 0.03085417, 0.07047743, 0.05618707, 0.0776226 ,
0.06138357, 0.07080221, 0.04124716, 0.04546931, 0.0659305 ,
0.06690484, 0.05878532, 0.05488795, 0.05293927, 0.04352062,
0.03637545, 0.0591101 , 0.06495615, 0.05975966, 0.05521273,
0.05846054, 0.04481975, 0.04514453, 0.06235791, 0.07697304])# Test data looks like
scaled_data_Malaga_y[235]0.07340045469308218# Since the first 90th values does not have historic, the dataset has been reduced in 90 values
print(f'Longitud datos de entrenamiento con historico: {len(scaled_data_Malaga_y)}')Longitud datos de entrenamiento con historico: 476# we split data in train and test
# as in previous analysis, we are going to predict a maximum of 91 days
x_train = scaled_data_Malaga_x[0:len(scaled_data_Malaga_x)-91]
y_train = scaled_data_Malaga_y[0:len(scaled_data_Malaga_y)-91]
print(f'Cantidad datos de entrenamiento: x={len(x_train)} - y={len(y_train)}')
x_test = scaled_data_Malaga_x[len(scaled_data_Malaga_x)-90:len(scaled_data_Malaga_x)]
y_test = scaled_data_Malaga_y[len(scaled_data_Malaga_y)-90:len(scaled_data_Malaga_y)]
print(f'Cantidad datos de test: x={len(x_test)} - y={len(y_test)}')Cantidad datos de entrenamiento: x=385 - y=385
Cantidad datos de test: x=90 - y=90# Reshape the data to feed de recurrent network
x_train = np.reshape(x_train, (x_train.shape[0], x_train.shape[1], 1))
print("Train data shape:")
print(x_train.shape)
print(y_train.shape)
x_test = np.reshape(x_test, (x_test.shape[0], x_test.shape[1], 1))
print("Test data shape:")
print(x_test.shape)
print(y_test.shape)Train data shape:
(385, 90, 1)
(385,)
Test data shape:
(90, 90, 1)
(90,)# Configure / setup the neural network model - LSTM
# Build the model
print('Build model...')
model = Sequential()
# Model with Neurons
# Inputshape = neurons -> Timestamps
neurons= x_train.shape[1]
model.add(LSTM(90,
activation = 'relu',
return_sequences = True,
input_shape = (x_train.shape[1], 1)))
model.add(LSTM(50,
activation = 'relu',
return_sequences = True))
model.add(LSTM(25,
activation = 'relu',
return_sequences = False))
model.add(Dense(5, activation = 'relu'))
model.add(Dense(1))Build model...model.compile(optimizer='adam', loss='mean_squared_error')
model.summary()Model: "sequential_8"_________________________________________________________________ Layer (type) Output Shape Param # ================================================================= lstm_24 (LSTM) (None, 90, 90) 33120 lstm_25 (LSTM) (None, 90, 50) 28200 lstm_26 (LSTM) (None, 25) 7600 dense_16 (Dense) (None, 5) 130 dense_17 (Dense) (None, 1) 6 =================================================================Total params: 69,056Trainable params: 69,056Non-trainable params: 0_________________________________________________________________# Training the model
# fit network
history = model.fit(x_train,
y_train,
batch_size = 1000,
epochs = 50,
validation_data = (x_test, y_test),
verbose = 0)plt.plot(history.history['loss'], label='train')
plt.plot(history.history['val_loss'], label='test')
plt.legend()
plt.show() 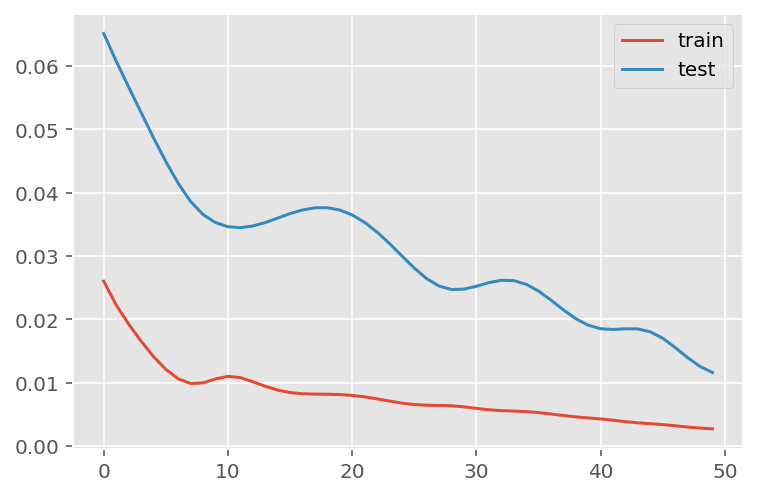
# Get the predicted values
predictions = model.predict(x_test)
predictions = scaler.inverse_transform(predictions)1/3 [=========>....................] - ETA: 0s3/3 [==============================] - ETA: 0s3/3 [==============================] - 0s 31ms/stepy_test = y_test.reshape(-1,1)
y_test = scaler.inverse_transform(y_test)# Calculate the mean absolute error (MAE)
mae = mean_absolute_error(y_test, predictions)
print('MAE: ' + str(round(mae, 1)))
# Calculate the root mean squarred error (RMSE)
rmse = np.sqrt(mean_squared_error(y_test,predictions))
print('RMSE: ' + str(round(rmse, 1)))
# Calculate the root mean squarred error (RMSE)
rmse = mean_squared_error(y_test,
predictions,
squared = False)
print('RMSE: ' + str(round(rmse, 1)))MAE: 197.7
RMSE: 331.7
RMSE: 331.7# Add the difference between the valid and predicted prices
train = data_Malaga[:(len(x_train)+92)]
valid = data_Malaga[(len(x_train)+91):]valid.insert(1, "Predictions", predictions, True)
valid.insert(1, "Difference", valid["Predictions"] - valid["num_casos"], True)# Zoom-in to a closer timeframe
# Date from which on the date is displayed
display_start_date = "2021-07-15"
valid = valid[valid.index > display_start_date]
train = train[train.index > display_start_date]# Visualize the data
matplotlib.style.use('ggplot')
fig, ax1 = plt.subplots(figsize=(22, 10), sharex=True)
# Data - Train
xt = train.index;
yt = train[["num_casos"]]
# Data - Test / validation
xv = valid.index;
yv = valid[["num_casos", "Predictions"]]
# Plot
plt.title("Malaga: Predictions vs Real infections", fontsize=20)
plt.ylabel("Nº Cases", fontsize=18)
plt.plot(yt, color="blue", linewidth=1.5)
plt.plot(yv["Predictions"], color="red", linewidth=1.5)
plt.plot(yv["num_casos"], color="green", linewidth=1.5)
plt.legend(["Train", "LSTM Predictions", "Test"],
loc="upper left", fontsize=18)
# Bar plot with the differences
x = valid.index
y = valid["Difference"]
plt.bar(x, y, width=0.2, color="grey")
plt.grid()
plt.show()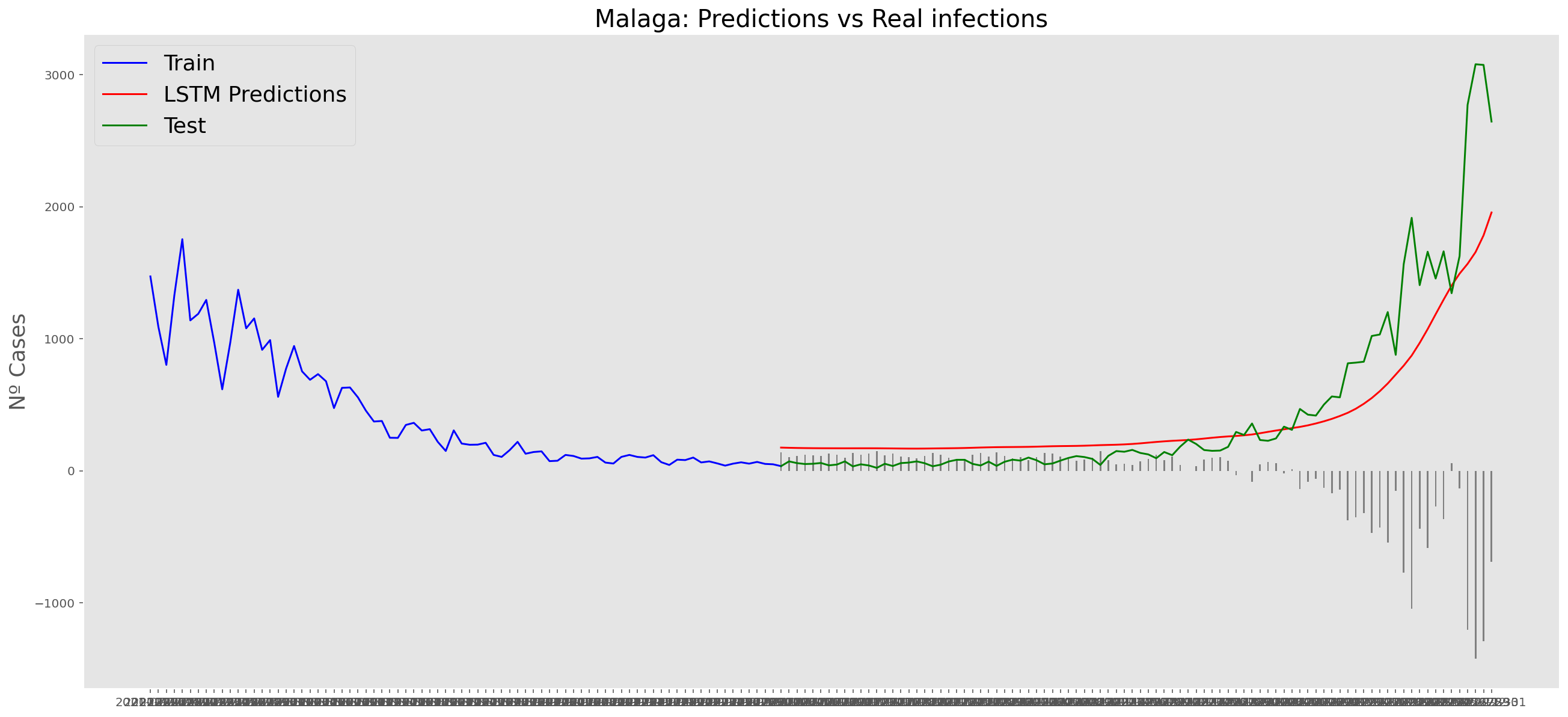
Sevilla
data_Sevilla = data_covid.loc[data_covid['provincia'] == 'Sevilla']
data_Sevilla = data_Sevilla.set_index('fecha')
data_Sevilla = data_Sevilla.filter(['num_casos'])
data_Sevilla = data_Sevilla['2020-06-14':'2021-12-31']
data_Sevilla| num_casos | |
|---|---|
| fecha | |
| 2020-06-14 | 0 |
| 2020-06-15 | 2 |
| 2020-06-16 | 1 |
| 2020-06-17 | 0 |
| 2020-06-18 | 0 |
| ... | ... |
| 2021-12-27 | 2617 |
| 2021-12-28 | 3190 |
| 2021-12-29 | 3692 |
| 2021-12-30 | 3508 |
| 2021-12-31 | 2816 |
566 rows × 1 columns
data_Sevilla.describe()| num_casos | |
|---|---|
| count | 566.000000 |
| mean | 383.807420 |
| std | 454.714408 |
| min | 0.000000 |
| 25% | 112.750000 |
| 50% | 260.500000 |
| 75% | 459.000000 |
| max | 3692.000000 |
np_data_Sevilla = data_Sevilla.valuesscaler = MinMaxScaler(feature_range=(0, 1))
scaled_data_Sevilla = scaler.fit_transform(np_data_Sevilla)
print(f'Longitud del conjunto de datos disponible: {len(scaled_data_Sevilla)}')Longitud del conjunto de datos disponible: 566# Since we are going to predict future values based on the X past elements,
# we need to create a list with those historic information for each element
historic_values = 90
scaled_data_Sevilla_x = []
scaled_data_Sevilla_y = []
for num_casos_i in range(historic_values, len(scaled_data_Sevilla)):
scaled_data_Sevilla_x.append(scaled_data_Sevilla[(num_casos_i-historic_values):num_casos_i, 0])
scaled_data_Sevilla_y.append(scaled_data_Sevilla[num_casos_i, 0])
# Convert the x_train and y_train to numpy arrays
scaled_data_Sevilla_x = np.array(scaled_data_Sevilla_x)
scaled_data_Sevilla_y = np.array(scaled_data_Sevilla_y)# Train data looks like
scaled_data_Sevilla_x[235]array([0.18634886, 0.19799567, 0.14626219, 0.0896533 , 0.13894908,
0.1695558 , 0.14572048, 0.12242687, 0.13434453, 0.06500542,
0.05390033, 0.08206934, 0.08667389, 0.09452871, 0.06798483,
0.05904659, 0.04252438, 0.0343987 , 0.06554713, 0.06473456,
0.05444204, 0.0503792 , 0.05796316, 0.04577465, 0.03819068,
0.04712893, 0.0528169 , 0.0663597 , 0.06013001, 0.05119177,
0.03764897, 0.031961 , 0.05119177, 0.05010834, 0.04739978,
0.05119177, 0.05390033, 0.03927411, 0.03737811, 0.06013001,
0.05633803, 0.05606717, 0.05850488, 0.06473456, 0.04658722,
0.04198267, 0.07150596, 0.07340195, 0.05823402, 0.08071506,
0.07773564, 0.05877573, 0.05471289, 0.07990249, 0.08829902,
0.0872156 , 0.09019502, 0.07069339, 0.08775731, 0.08396533,
0.14951246, 0.12432286, 0.12269772, 0.12378115, 0.13028169,
0.11348862, 0.07936078, 0.13299025, 0.12269772, 0.11213434,
0.12107259, 0.12432286, 0.09723727, 0.07042254, 0.09886241,
0.11782232, 0.10157096, 0.10346696, 0.12621885, 0.08342362,
0.07340195, 0.0847779 , 0.08559047, 0.11159263, 0.10292524,
0.08911159, 0.07502709, 0.04821235, 0.08315276, 0.09913326])# Test data looks like
scaled_data_Sevilla_y[235]0.10861321776814734# Since the first 90th values does not have historic, the dataset has been reduced in 90 values
print(f'Longitud datos de entrenamiento con historico: {len(scaled_data_Sevilla_y)}')Longitud datos de entrenamiento con historico: 476# we split data in train and test
# as in previous analysis, we are going to predict a maximum of 91 days
x_train = scaled_data_Sevilla_x[0:len(scaled_data_Sevilla_x)-91]
y_train = scaled_data_Sevilla_y[0:len(scaled_data_Sevilla_y)-91]
print(f'Cantidad datos de entrenamiento: x={len(x_train)} - y={len(y_train)}')
x_test = scaled_data_Sevilla_x[len(scaled_data_Sevilla_x)-90:len(scaled_data_Sevilla_x)]
y_test = scaled_data_Sevilla_y[len(scaled_data_Sevilla_y)-90:len(scaled_data_Sevilla_y)]
print(f'Cantidad datos de test: x={len(x_test)} - y={len(y_test)}')Cantidad datos de entrenamiento: x=385 - y=385
Cantidad datos de test: x=90 - y=90# Reshape the data to feed de recurrent network
x_train = np.reshape(x_train, (x_train.shape[0], x_train.shape[1], 1))
print("Train data shape:")
print(x_train.shape)
print(y_train.shape)
x_test = np.reshape(x_test, (x_test.shape[0], x_test.shape[1], 1))
print("Test data shape:")
print(x_test.shape)
print(y_test.shape)Train data shape:
(385, 90, 1)
(385,)
Test data shape:
(90, 90, 1)
(90,)# Configure / setup the neural network model - LSTM
# Build the model
print('Build model...')
model = Sequential()
# Model with Neurons
# Inputshape = neurons -> Timestamps
neurons= x_train.shape[1]
model.add(LSTM(90,
activation = 'relu',
return_sequences = True,
input_shape = (x_train.shape[1], 1)))
model.add(LSTM(50,
activation = 'relu',
return_sequences = True))
model.add(LSTM(25,
activation = 'relu',
return_sequences = False))
model.add(Dense(5, activation = 'relu'))
model.add(Dense(1))Build model...model.compile(optimizer='adam', loss='mean_squared_error')
model.summary()Model: "sequential_9"_________________________________________________________________ Layer (type) Output Shape Param # ================================================================= lstm_27 (LSTM) (None, 90, 90) 33120 lstm_28 (LSTM) (None, 90, 50) 28200 lstm_29 (LSTM) (None, 25) 7600 dense_18 (Dense) (None, 5) 130 dense_19 (Dense) (None, 1) 6 =================================================================Total params: 69,056Trainable params: 69,056Non-trainable params: 0_________________________________________________________________# Training the model
# fit network
history = model.fit(x_train,
y_train,
batch_size = 1000,
epochs = 50,
validation_data = (x_test, y_test),
verbose = 0)plt.plot(history.history['loss'], label='train')
plt.plot(history.history['val_loss'], label='test')
plt.legend()
plt.show() 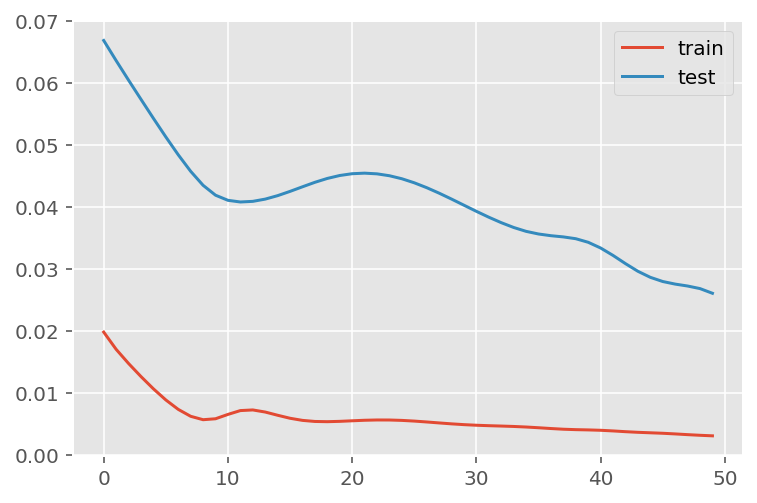
# Get the predicted values
predictions = model.predict(x_test)
predictions = scaler.inverse_transform(predictions)1/3 [=========>....................] - ETA: 0s3/3 [==============================] - ETA: 0s3/3 [==============================] - 0s 32ms/stepy_test = y_test.reshape(-1,1)
y_test = scaler.inverse_transform(y_test)# Calculate the mean absolute error (MAE)
mae = mean_absolute_error(y_test, predictions)
print('MAE: ' + str(round(mae, 1)))
# Calculate the root mean squarred error (RMSE)
rmse = np.sqrt(mean_squared_error(y_test,predictions))
print('RMSE: ' + str(round(rmse, 1)))
# Calculate the root mean squarred error (RMSE)
rmse = mean_squared_error(y_test,
predictions,
squared = False)
print('RMSE: ' + str(round(rmse, 1)))MAE: 329.2
RMSE: 595.9
RMSE: 595.9# Add the difference between the valid and predicted prices
train = data_Sevilla[:(len(x_train)+92)]
valid = data_Sevilla[(len(x_train)+91):]valid.insert(1, "Predictions", predictions, True)
valid.insert(1, "Difference", valid["Predictions"] - valid["num_casos"], True)# Zoom-in to a closer timeframe
# Date from which on the date is displayed
display_start_date = "2021-07-15"
valid = valid[valid.index > display_start_date]
train = train[train.index > display_start_date]# Visualize the data
matplotlib.style.use('ggplot')
fig, ax1 = plt.subplots(figsize=(22, 10), sharex=True)
# Data - Train
xt = train.index;
yt = train[["num_casos"]]
# Data - Test / validation
xv = valid.index;
yv = valid[["num_casos", "Predictions"]]
# Plot
plt.title("Sevilla: Predictions vs Real infections", fontsize=20)
plt.ylabel("Nº Cases", fontsize=18)
plt.plot(yt, color="blue", linewidth=1.5)
plt.plot(yv["Predictions"], color="red", linewidth=1.5)
plt.plot(yv["num_casos"], color="green", linewidth=1.5)
plt.legend(["Train", "LSTM Predictions", "Test"],
loc="upper left", fontsize=18)
# Bar plot with the differences
x = valid.index
y = valid["Difference"]
plt.bar(x, y, width=0.2, color="grey")
plt.grid()
plt.show()