pacman::p_load(
here, # file locator
tidyverse, # data management and ggplot2 graphics
skimr, # get overview of data
janitor, # produce and adorn tabulations and cross-tabulations
tsibble,
fable,
feasts,
fabletools,
lubridate
)
covid_data <- readRDS(here("data", "clean", "final_covid_data.rds"))Multivariate (Google mobility + temp) prediction
Load packages:
The temporal limits of our prediction will be:
- Data as of June 14th 2020 which is considered the beginning of the second epidemiological period in Spain
- In the first case scenario, data previous the start of the sixth wave of covid-19 (14 oct 2021).
- In the second case scenario, all the available data (limited by mobility information 29 dec 2021)
Before sixth epidemiological period
Asturias
data_asturias <- covid_data %>%
filter(provincia == "Asturias") %>%
filter(fecha > as.Date("2020-06-14", format = "%Y-%m-%d")) %>%
select(provincia, fecha, num_casos, tmed, mob_grocery_pharmacy, mob_parks, mob_residential, mob_retail_recreation, mob_transit_stations, mob_workplaces) %>%
drop_na() %>%
as_tsibble(key = provincia, index = fecha)
data_asturias# A tsibble: 653 x 10 [1D]
# Key: provincia [1]
provincia fecha num_casos tmed mob_grocery_pharmacy mob_parks
<chr> <date> <dbl> <dbl> <dbl> <dbl>
1 Asturias 2020-06-15 0 16 -6 39
2 Asturias 2020-06-16 1 15.6 -6 7
3 Asturias 2020-06-17 0 15.6 -7 20
4 Asturias 2020-06-18 0 15.1 -4 68
5 Asturias 2020-06-19 0 14.8 -4 52
6 Asturias 2020-06-20 0 16.8 -10 71
7 Asturias 2020-06-21 0 18.2 -5.5 46
8 Asturias 2020-06-22 0 18.2 -1 99
9 Asturias 2020-06-23 0 20 -2 129
10 Asturias 2020-06-24 0 18.2 -9 63
# … with 643 more rows, and 4 more variables: mob_residential <dbl>,
# mob_retail_recreation <dbl>, mob_transit_stations <dbl>,
# mob_workplaces <dbl>data_asturias_train <- data_asturias %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d"))
summary(data_asturias_train$fecha) Min. 1st Qu. Median Mean 3rd Qu. Max.
"2020-06-15" "2020-10-14" "2021-02-13" "2021-02-13" "2021-06-14" "2021-10-14" data_asturias_test <- data_asturias %>%
filter(fecha > as.Date("2021-10-14", format = "%Y-%m-%d"))
summary(data_asturias_test$fecha) Min. 1st Qu. Median Mean 3rd Qu. Max.
"2021-10-15" "2021-11-25" "2022-01-05" "2022-01-05" "2022-02-15" "2022-03-29" # Lamba for num_casos
lambda_asturias_casos <- data_asturias %>%
features(num_casos, features = guerrero) %>%
pull(lambda_guerrero)
lambda_asturias_casos[1] 0.2278229# Lamba for average temp
lambda_asturias_tmed <- data_asturias %>%
features(tmed, features = guerrero) %>%
pull(lambda_guerrero)
lambda_asturias_tmed[1] 1.056074# Lamba for mobility
lambda_asturias_grocery <- data_asturias %>%
features(mob_grocery_pharmacy, features = guerrero) %>%
pull(lambda_guerrero)
lambda_asturias_grocery[1] 1.377952lambda_asturias_parks <- data_asturias %>%
features(mob_parks, features = guerrero) %>%
pull(lambda_guerrero)
lambda_asturias_parks[1] 0.7188469lambda_asturias_resd <- data_asturias %>%
features(mob_residential, features = guerrero) %>%
pull(lambda_guerrero)
lambda_asturias_resd[1] 1.000064lambda_asturias_retail <- data_asturias %>%
features(mob_retail_recreation, features = guerrero) %>%
pull(lambda_guerrero)
lambda_asturias_retail[1] 1.058286lambda_asturias_transit <- data_asturias %>%
features(mob_transit_stations, features = guerrero) %>%
pull(lambda_guerrero)
lambda_asturias_transit[1] 1.116829lambda_asturias_work <- data_asturias %>%
features(mob_workplaces, features = guerrero) %>%
pull(lambda_guerrero)
lambda_asturias_work[1] 1.999927fit_model <- data_asturias_train %>%
model(
arima_at1 = ARIMA(box_cox(num_casos, lambda_asturias_casos) ~
box_cox(tmed, lambda_asturias_tmed) +
box_cox(mob_grocery_pharmacy, lambda_asturias_grocery) +
box_cox(mob_parks, lambda_asturias_parks) +
box_cox(mob_residential, lambda_asturias_resd) +
box_cox(mob_retail_recreation, lambda_asturias_retail) +
box_cox(mob_transit_stations, lambda_asturias_transit) +
box_cox(mob_workplaces, lambda_asturias_work)
),
arima_at2 = ARIMA(box_cox(num_casos, lambda_asturias_casos) ~
box_cox(tmed, lambda_asturias_tmed) +
box_cox(mob_grocery_pharmacy, lambda_asturias_grocery) +
box_cox(mob_parks, lambda_asturias_parks) +
box_cox(mob_residential, lambda_asturias_resd) +
box_cox(mob_retail_recreation, lambda_asturias_retail) +
box_cox(mob_transit_stations, lambda_asturias_transit) +
box_cox(mob_workplaces, lambda_asturias_work),
stepwise = FALSE, approx = FALSE),
Snaive = SNAIVE(box_cox(num_casos, lambda_asturias_casos))
)Warning in sqrt(diag(best$var.coef)): NaNs produced# Show and report model
fit_model# A mable: 1 x 4
# Key: provincia [1]
provincia arima_at1
<chr> <model>
1 Asturias <LM w/ ARIMA(4,0,0)(2,0,0)[7] errors>
# … with 2 more variables: arima_at2 <model>, Snaive <model>glance(fit_model)# A tibble: 3 × 9
provincia .model sigma2 log_lik AIC AICc BIC ar_roots ma_roots
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <list> <list>
1 Asturias arima_at1 0.955 -675. 1380. 1381. 1443. <cpl [18]> <cpl [0]>
2 Asturias arima_at2 0.922 -668. 1363. 1364. 1417. <cpl [9]> <cpl [8]>
3 Asturias Snaive 2.32 NA NA NA NA <NULL> <NULL> fit_model %>%
pivot_longer(-provincia,
names_to = ".model",
values_to = "orders") %>%
left_join(glance(fit_model), by = c("provincia", ".model")) %>%
arrange(AICc) %>%
select(.model:BIC)# A mable: 3 x 7
# Key: .model [3]
.model orders sigma2 log_lik AIC AICc BIC
<chr> <model> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_… <LM w/ ARIMA(2,0,1)(1,0,1)[7] errors> 0.922 -668. 1363. 1364. 1417.
2 arima_… <LM w/ ARIMA(4,0,0)(2,0,0)[7] errors> 0.955 -675. 1380. 1381. 1443.
3 Snaive <SNAIVE> 2.32 NA NA NA NA # We use a Ljung-Box test >> large p-value, confirms residuals are similar / considered to white noise
fit_model %>%
select(Snaive) %>%
gg_tsresiduals()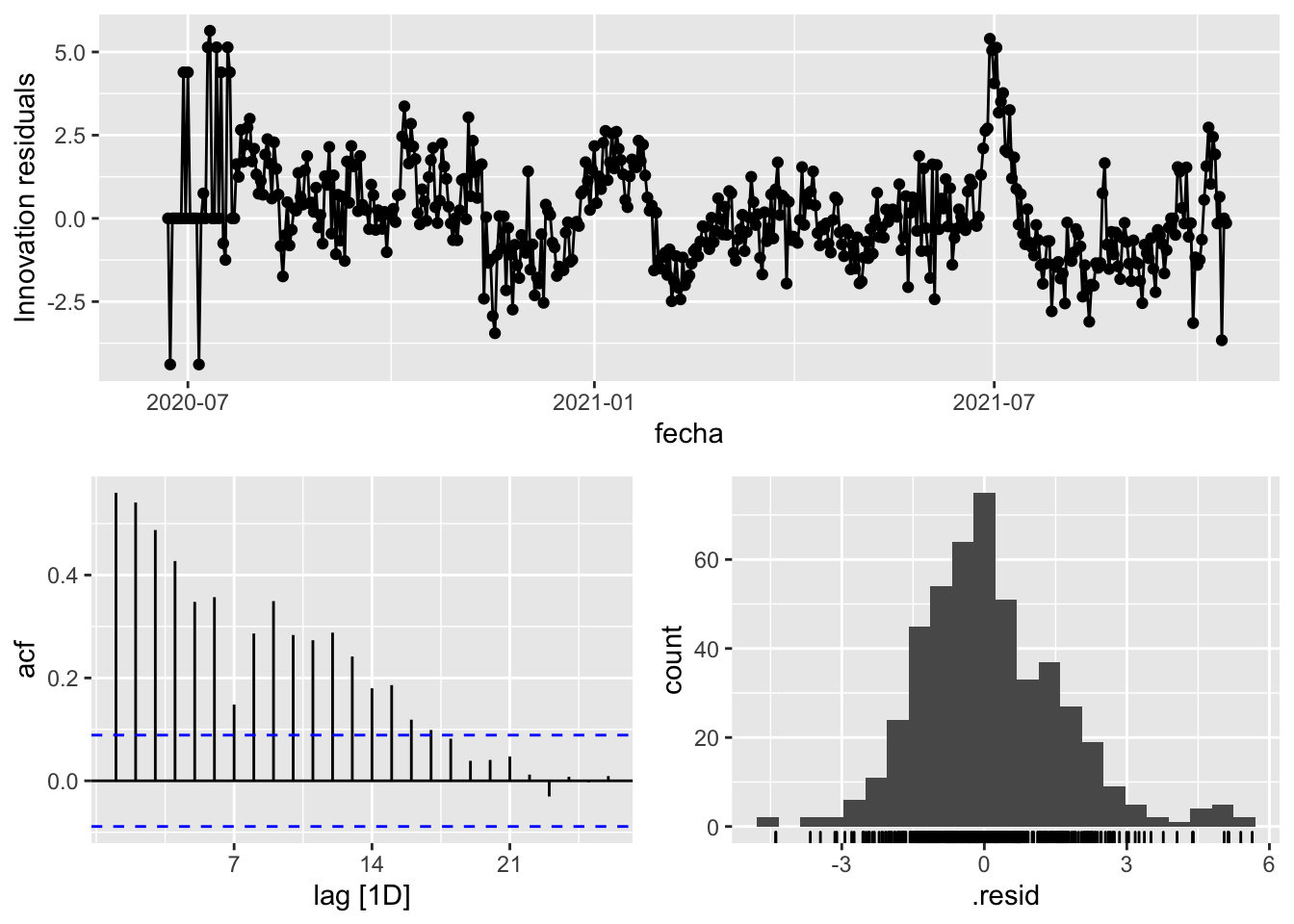
fit_model %>%
select(arima_at1) %>%
gg_tsresiduals()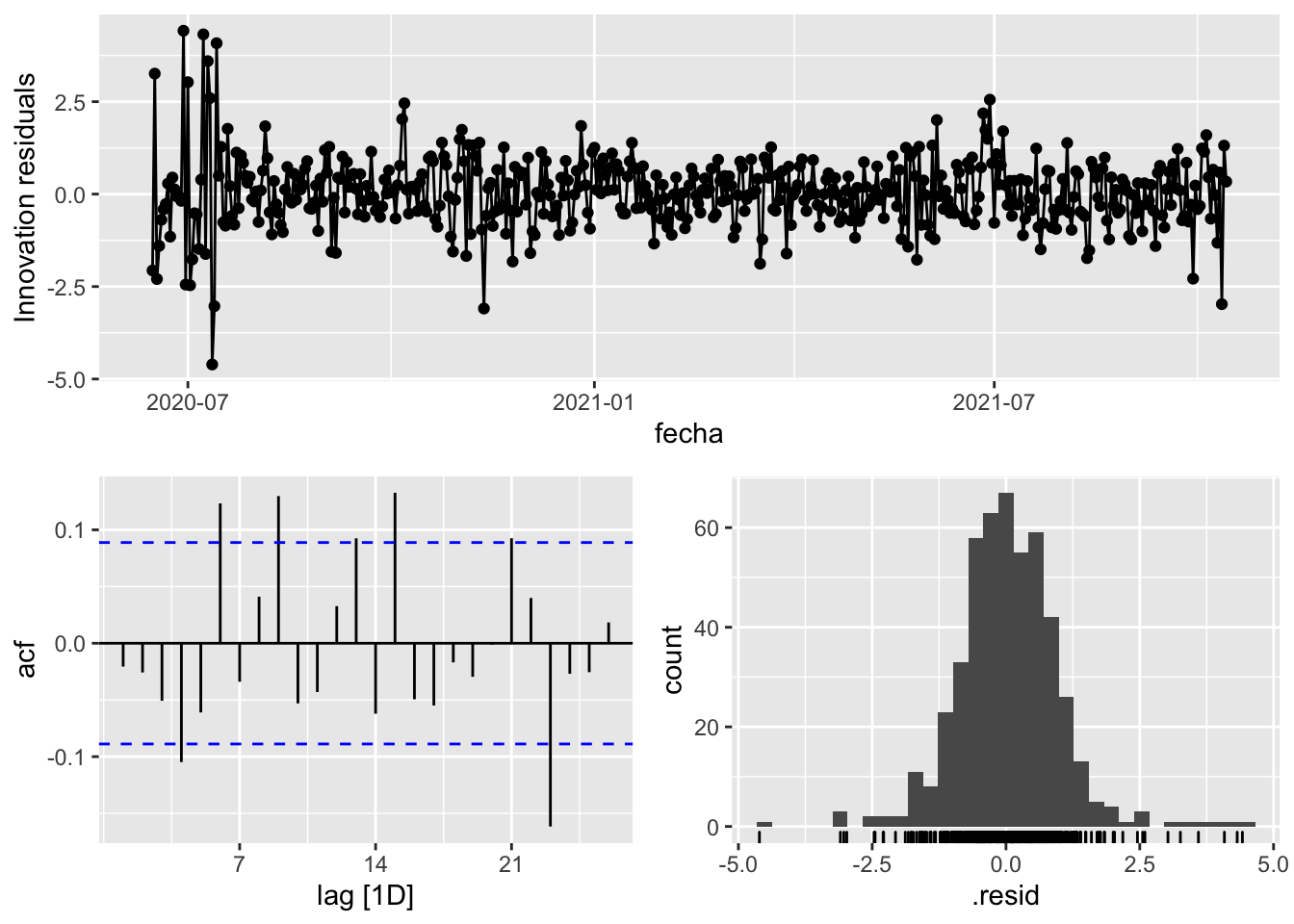
fit_model %>%
select(arima_at2) %>%
gg_tsresiduals()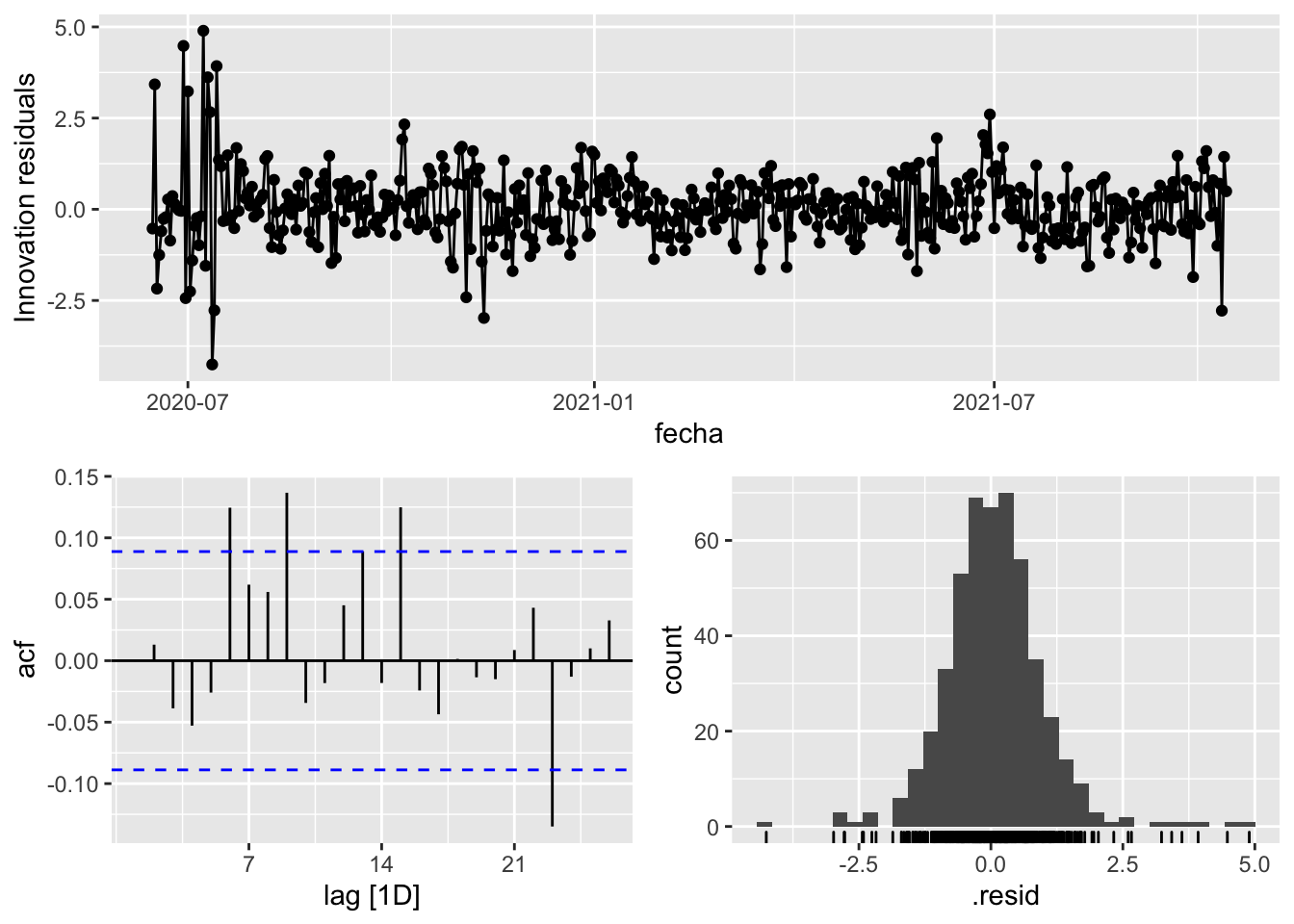
augment(fit_model) %>%
features(.innov, ljung_box, lag=7)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Asturias arima_at1 17.1 0.0165
2 Asturias arima_at2 12.1 0.0972
3 Asturias Snaive 629. 0 augment(fit_model) %>%
features(.innov, ljung_box, lag=14)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Asturias arima_at1 35.5 0.00125
2 Asturias arima_at2 28.9 0.0108
3 Asturias Snaive 892. 0 augment(fit_model) %>%
features(.innov, ljung_box, lag=21)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Asturias arima_at1 52.1 0.000185
2 Asturias arima_at2 38.2 0.0121
3 Asturias Snaive 927. 0 augment(fit_model) %>%
features(.innov, ljung_box, lag=90)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Asturias arima_at1 128. 0.00526
2 Asturias arima_at2 99.4 0.233
3 Asturias Snaive 1195. 0 # To predict future values of infections, we need to pass the model future/prediction values of the complementary variables tmed and mobility
# We are going to select those from the test dataset
data_fc_h7 <- data_asturias_test %>%
select(-num_casos) %>%
slice(1:7)
data_fc_h14 <- data_asturias_test %>%
select(-num_casos) %>%
slice(1:14)
data_fc_h21 <- data_asturias_test %>%
select(-num_casos) %>%
slice(1:21)
data_fc_h90 <- data_asturias_test %>%
select(-num_casos) %>%
slice(1:90)# Significant spikes out of 30 is still consistent with white noise
# To be sure, use a Ljung-Box test, which has a large p-value, confirming that
# the residuals are similar to white noise.
# Note that the alternative models also passed this test.
#
# Forecast
fc_h7 <- fabletools::forecast(fit_model, new_data = data_fc_h7)
fc_h14 <- fabletools::forecast(fit_model, new_data = data_fc_h14)
fc_h21 <- fabletools::forecast(fit_model, new_data = data_fc_h21)
fc_h90 <- fabletools::forecast(fit_model, new_data = data_fc_h90)7 days
# Plots
fc_h7 %>%
autoplot(
data_asturias_test %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(7))
) +
labs(y = "Nº cases", title = "Asturias - forecast h7")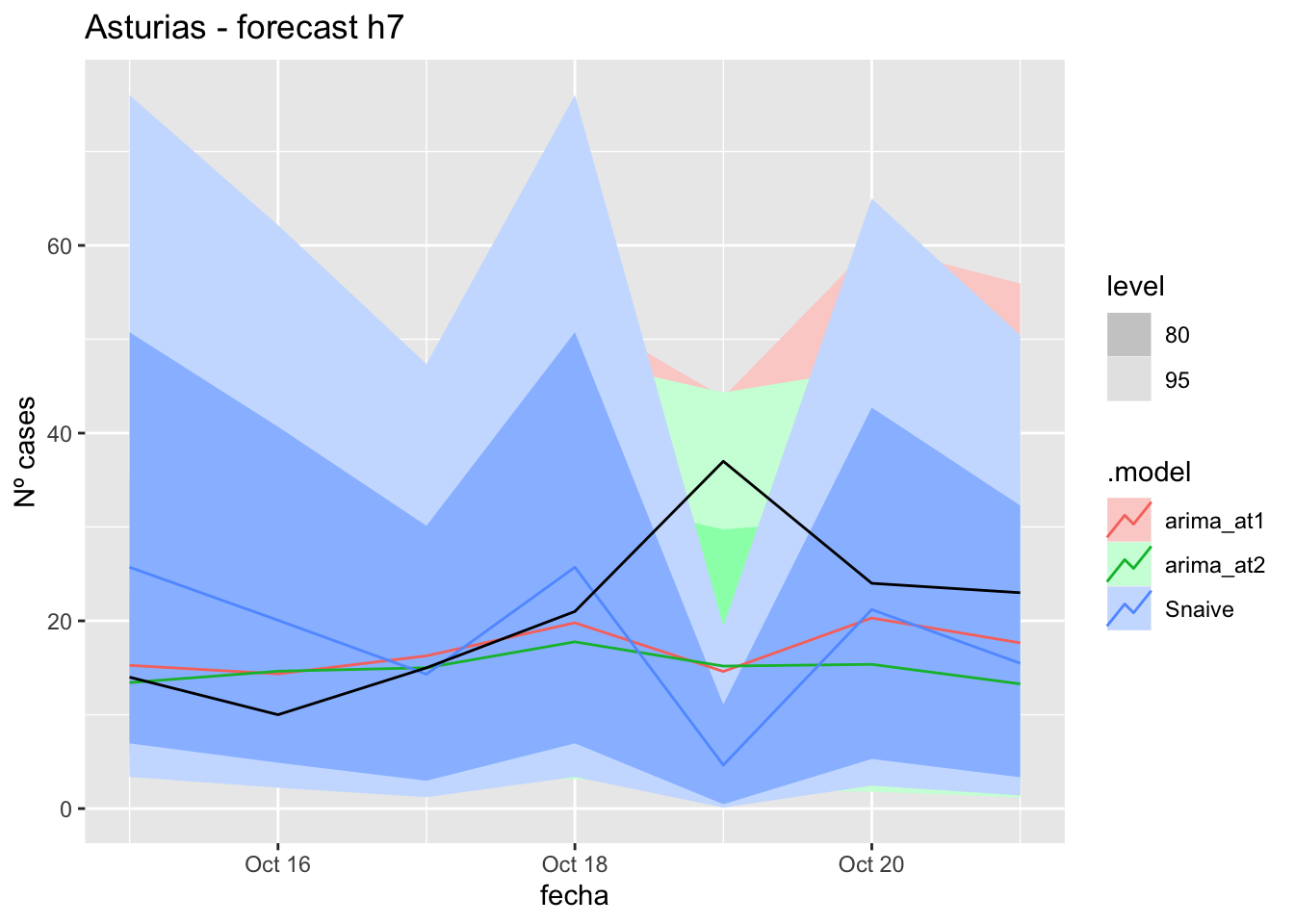
fc_h7 %>%
autoplot(
data_asturias %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(7) &
fecha > as.Date("2021-07-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Asturias - forecast h7")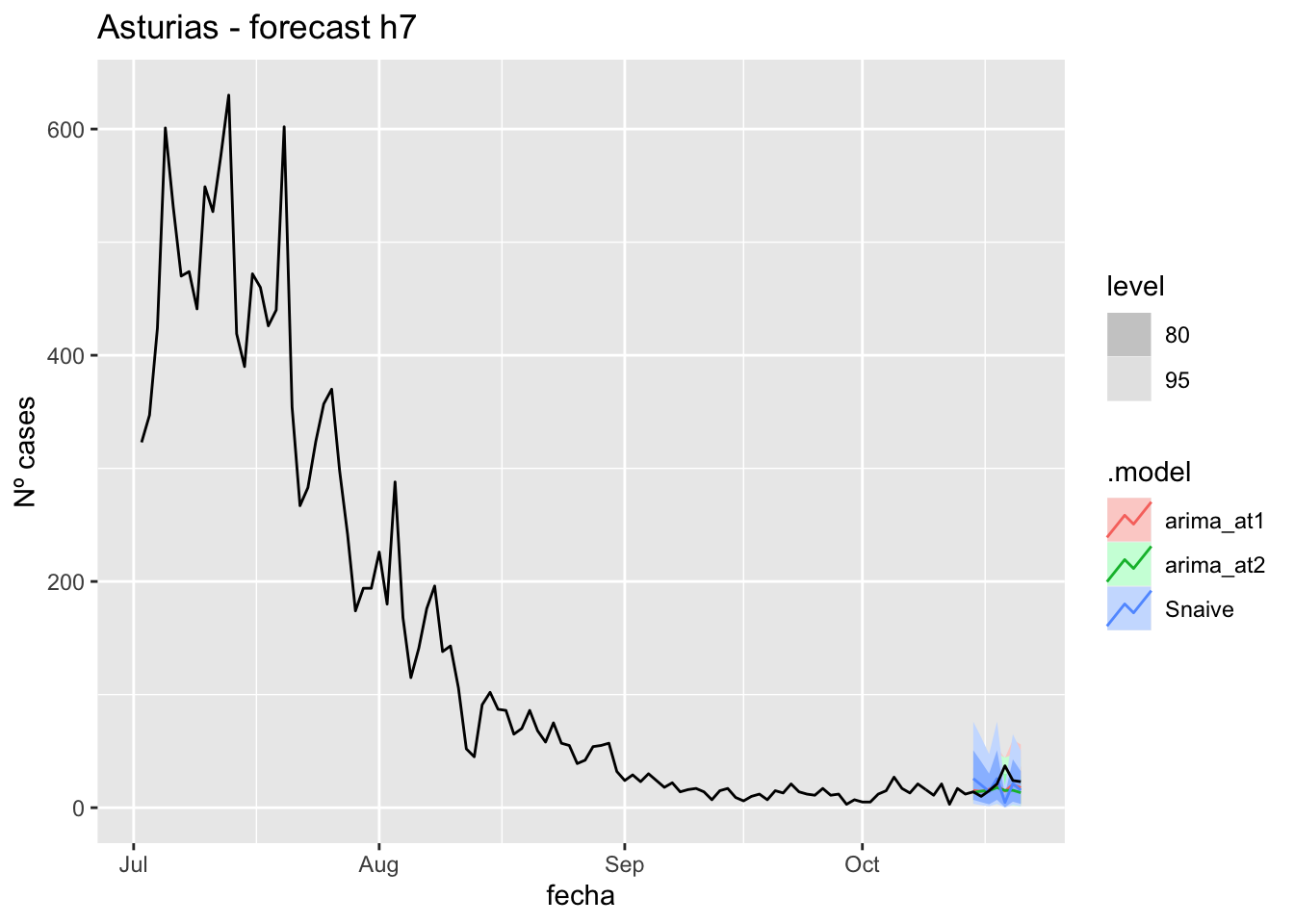
# Accuracy
fabletools::accuracy(fc_h7, data_asturias)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_at1 Asturias Test 3.68 9.00 5.65 6.27 23.7 0.114 0.110 0.0968
2 arima_at2 Asturias Test 5.62 9.84 6.95 15.8 29.0 0.140 0.120 0.318
3 Snaive Asturias Test 2.41 14.0 9.98 -10.1 49.0 0.201 0.171 0.0076614 days
# Plots
fc_h14 %>%
autoplot(
data_asturias_test %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(14))
) +
labs(y = "Nº cases", title = "Asturias - forecast h14")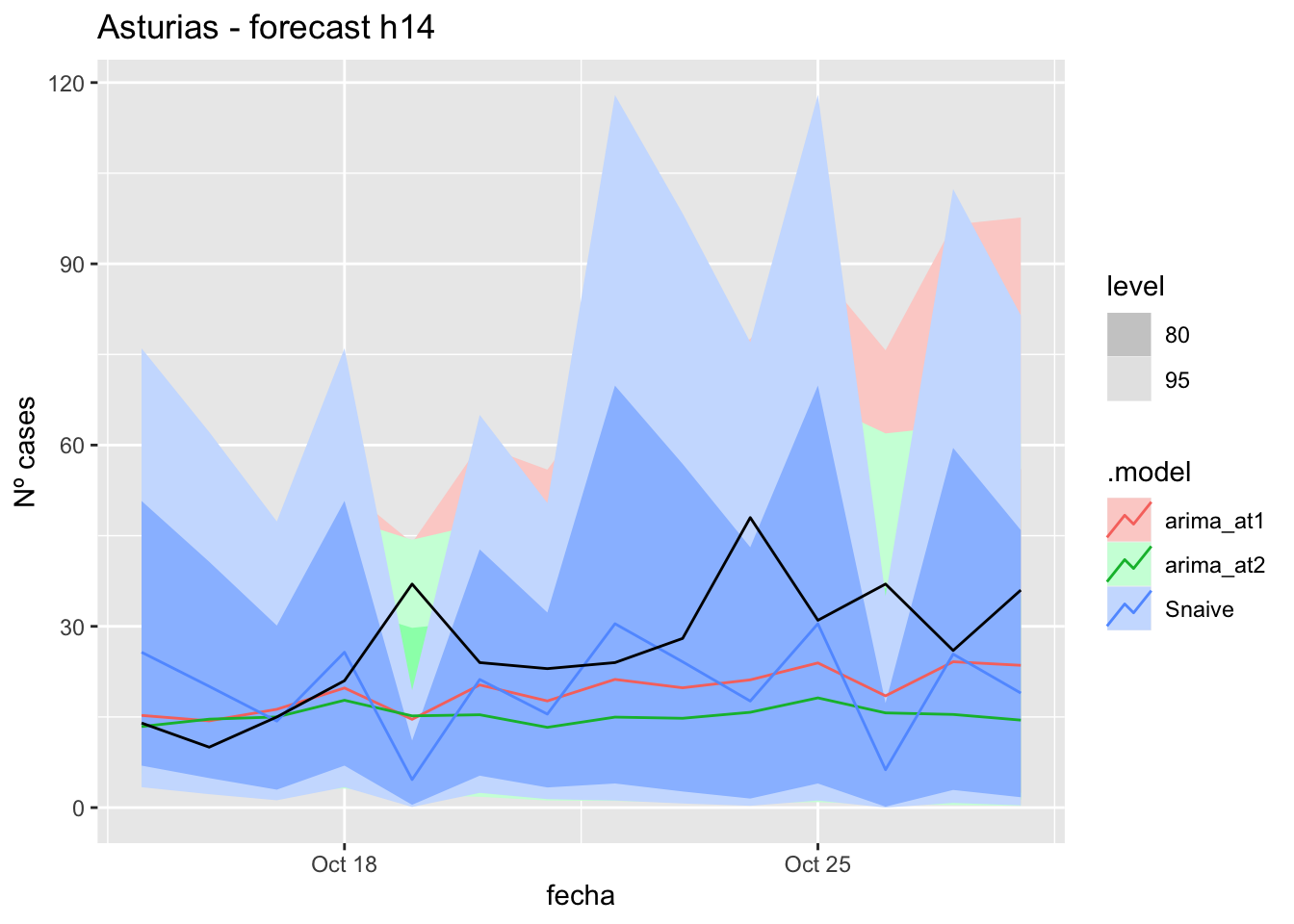
fc_h14 %>%
autoplot(
data_asturias %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(14) &
fecha > as.Date("2021-07-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Asturias - forecast h14")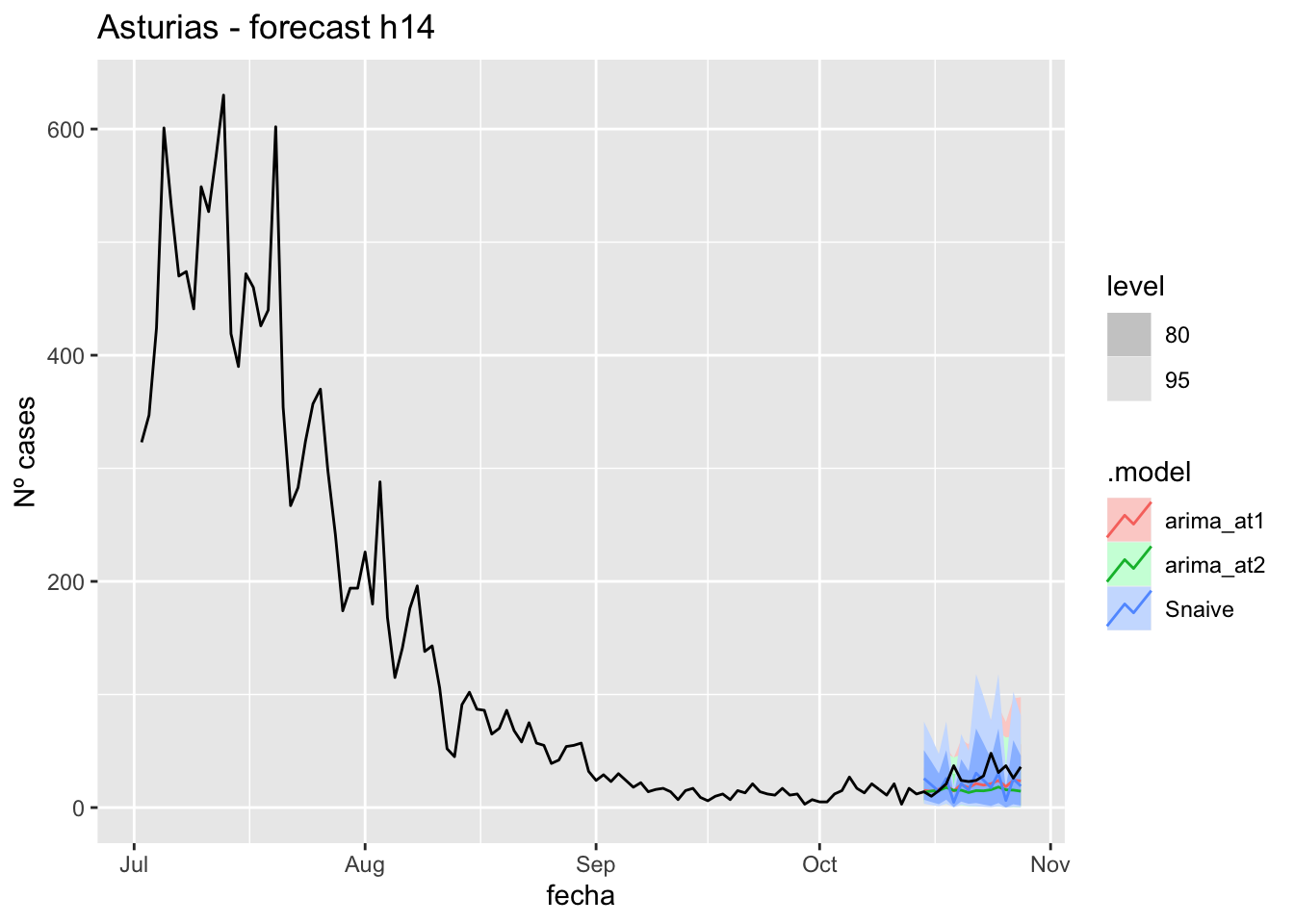
# Accuracy
fabletools::accuracy(fc_h14, data_asturias)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_at1 Asturias Test 7.39 11.7 8.37 18.2 26.9 0.168 0.142 0.0327
2 arima_at2 Asturias Test 11.4 15.0 12.1 33.0 39.6 0.243 0.183 0.304
3 Snaive Asturias Test 6.69 16.0 11.4 8.17 41.5 0.229 0.195 -0.156 21 days
# Plots
fc_h21 %>%
autoplot(
data_asturias_test %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(21))
) +
labs(y = "Nº cases", title = "Asturias - forecast h21")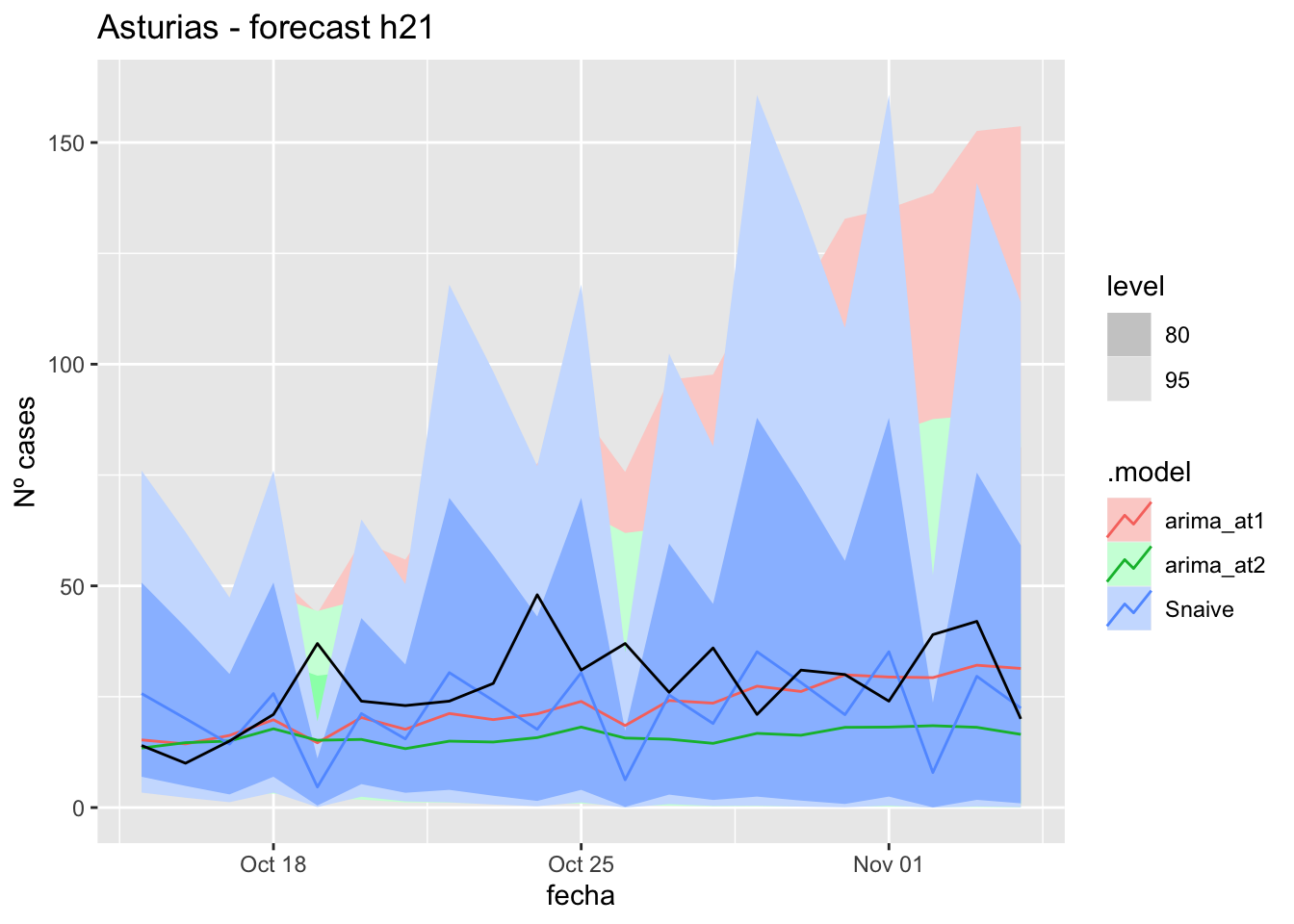
fc_h21 %>%
autoplot(
data_asturias %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(21) &
fecha > as.Date("2021-07-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Asturias - forecast h21")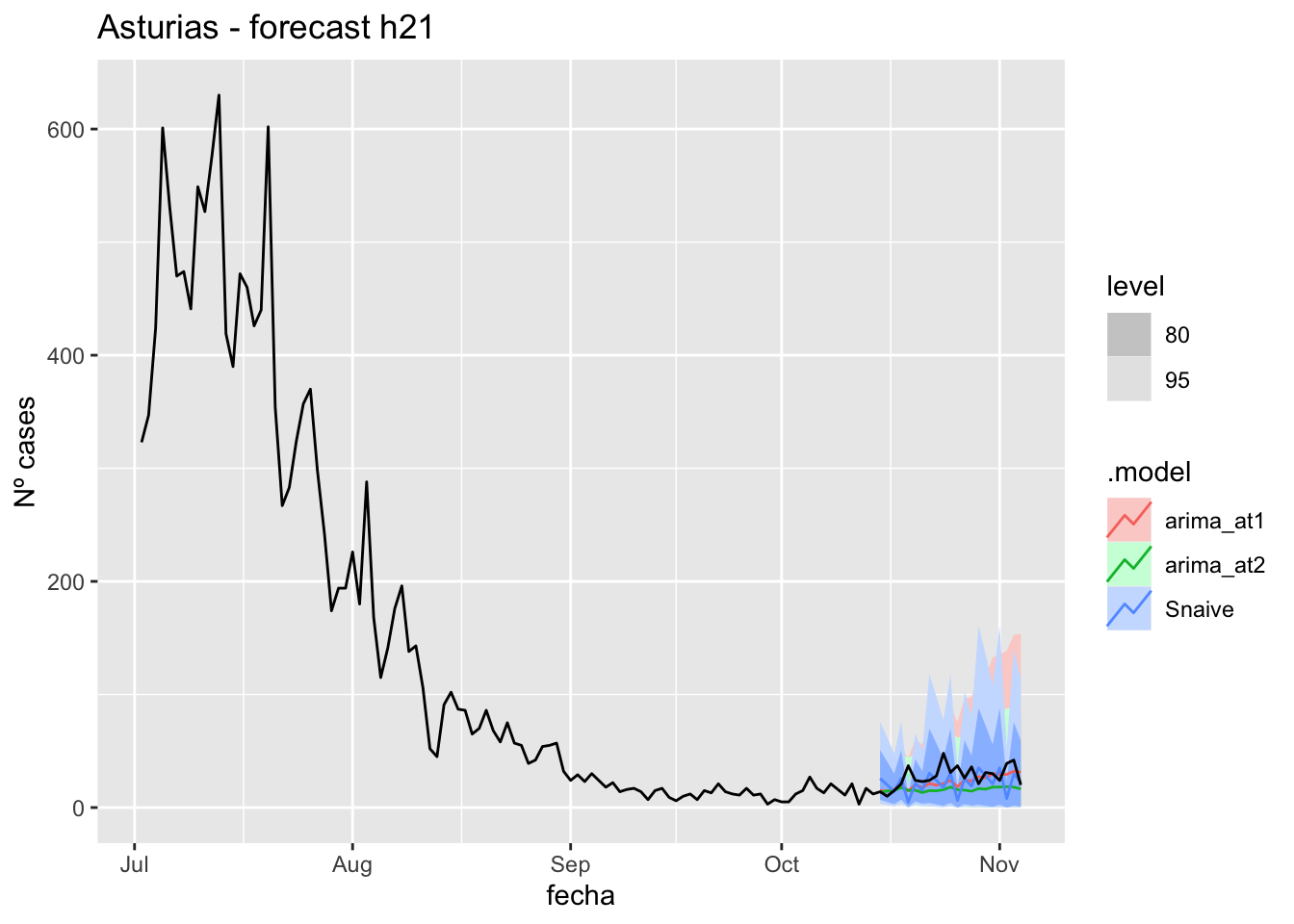
# Accuracy
fabletools::accuracy(fc_h21, data_asturias)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_at1 Asturias Test 4.99 10.5 7.85 9.97 26.3 0.158 0.128 -0.00881
2 arima_at2 Asturias Test 11.7 14.8 12.1 34.3 38.7 0.243 0.180 0.155
3 Snaive Asturias Test 5.77 15.6 11.5 6.52 40.8 0.232 0.190 -0.223 90 days
# Plots
fc_h90 %>%
autoplot(
data_asturias_test %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(90))
) +
labs(y = "Nº cases", title = "Asturias - forecast h90")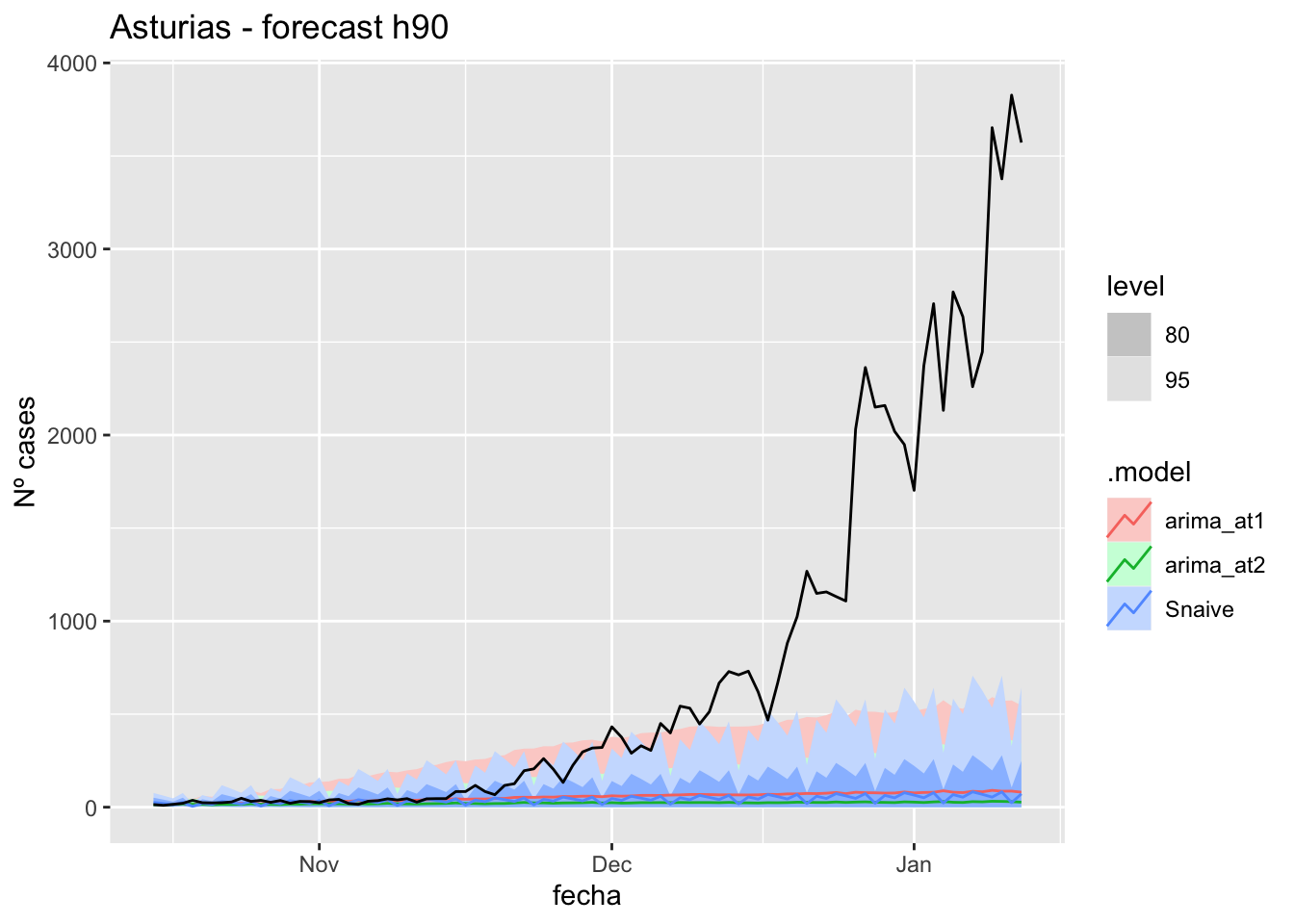
fc_h90 %>%
autoplot(
data_asturias %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(90) &
fecha > as.Date("2021-07-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Asturias - forecast h90")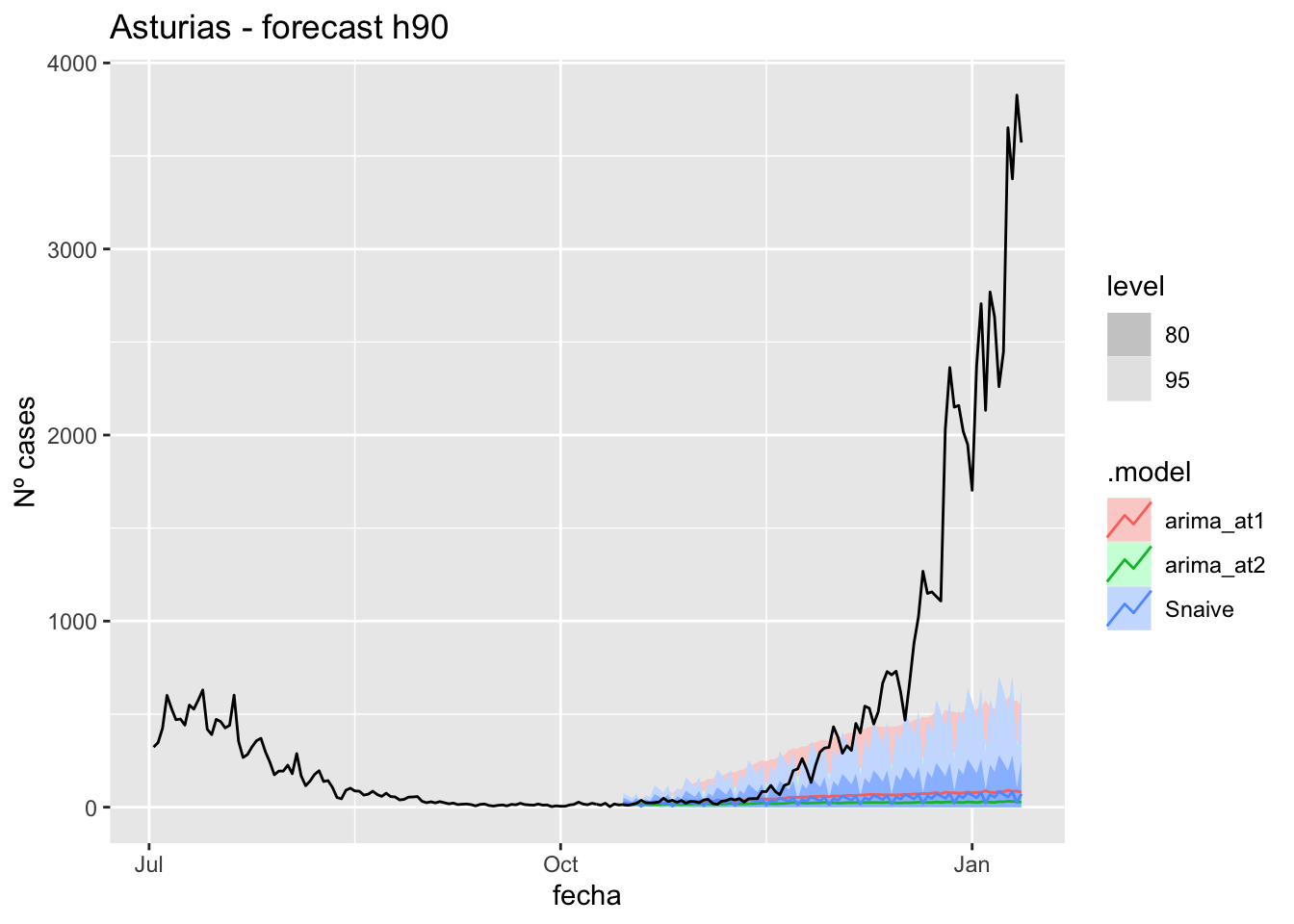
# Accuracy
fabletools::accuracy(fc_h90, data_asturias)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_at1 Asturias Test 689. 1200. 691. 56.5 64.2 13.9 14.6 0.925
2 arima_at2 Asturias Test 721. 1229. 721. 74.4 75.9 14.5 15.0 0.926
3 Snaive Asturias Test 702. 1213. 704. 60.0 71.7 14.2 14.8 0.925Barcelona
data_Barcelona <- covid_data %>%
filter(provincia == "Barcelona") %>%
filter(fecha > as.Date("2020-06-14", format = "%Y-%m-%d")) %>%
select(provincia, fecha, num_casos, tmed, mob_grocery_pharmacy, mob_parks, mob_residential, mob_retail_recreation, mob_transit_stations, mob_workplaces) %>%
drop_na() %>%
as_tsibble(key = provincia, index = fecha)
data_Barcelona# A tsibble: 653 x 10 [1D]
# Key: provincia [1]
provincia fecha num_casos tmed mob_grocery_pharmacy mob_parks
<chr> <date> <dbl> <dbl> <dbl> <dbl>
1 Barcelona 2020-06-15 62 21.2 -9 17
2 Barcelona 2020-06-16 66 20.8 -10 -16
3 Barcelona 2020-06-17 70 20.3 -4 14
4 Barcelona 2020-06-18 68 18.8 -8 -6
5 Barcelona 2020-06-19 65 20.4 -5 10
6 Barcelona 2020-06-20 53 21.8 -12 15
7 Barcelona 2020-06-21 38 22.8 -18 18
8 Barcelona 2020-06-22 69 23.8 5 24
9 Barcelona 2020-06-23 95 23.4 19 28
10 Barcelona 2020-06-24 44 23.6 -71 28
# … with 643 more rows, and 4 more variables: mob_residential <dbl>,
# mob_retail_recreation <dbl>, mob_transit_stations <dbl>,
# mob_workplaces <dbl>data_Barcelona_train <- data_Barcelona %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d"))
summary(data_Barcelona_train$fecha) Min. 1st Qu. Median Mean 3rd Qu. Max.
"2020-06-15" "2020-10-14" "2021-02-13" "2021-02-13" "2021-06-14" "2021-10-14" data_Barcelona_test <- data_Barcelona %>%
filter(fecha > as.Date("2021-10-14", format = "%Y-%m-%d"))
summary(data_Barcelona_test$fecha) Min. 1st Qu. Median Mean 3rd Qu. Max.
"2021-10-15" "2021-11-25" "2022-01-05" "2022-01-05" "2022-02-15" "2022-03-29" # Lamba for num_casos
lambda_Barcelona_casos <- data_asturias %>%
features(num_casos, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Barcelona_casos[1] 0.2278229# Lamba for average temp
lambda_Barcelona_tmed <- data_asturias %>%
features(tmed, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Barcelona_tmed[1] 1.056074# Lamba for mobility
lambda_Barcelona_grocery <- data_asturias %>%
features(mob_grocery_pharmacy, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Barcelona_grocery[1] 1.377952lambda_Barcelona_parks <- data_asturias %>%
features(mob_parks, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Barcelona_parks[1] 0.7188469lambda_Barcelona_resd <- data_asturias %>%
features(mob_residential, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Barcelona_resd[1] 1.000064lambda_Barcelona_retail <- data_asturias %>%
features(mob_retail_recreation, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Barcelona_retail[1] 1.058286lambda_Barcelona_transit <- data_asturias %>%
features(mob_transit_stations, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Barcelona_transit[1] 1.116829lambda_Barcelona_work <- data_asturias %>%
features(mob_workplaces, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Barcelona_work[1] 1.999927fit_model <- data_Barcelona_train %>%
model(
arima_at1 = ARIMA(box_cox(num_casos, lambda_Barcelona_casos) ~
box_cox(tmed, lambda_Barcelona_tmed) +
box_cox(mob_grocery_pharmacy, lambda_Barcelona_grocery) +
box_cox(mob_parks, lambda_Barcelona_parks) +
box_cox(mob_residential, lambda_Barcelona_resd) +
box_cox(mob_retail_recreation, lambda_Barcelona_retail) +
box_cox(mob_transit_stations, lambda_Barcelona_transit) +
box_cox(mob_workplaces, lambda_Barcelona_work)
),
arima_at2 = ARIMA(box_cox(num_casos, lambda_Barcelona_casos) ~
box_cox(tmed, lambda_Barcelona_tmed) +
box_cox(mob_grocery_pharmacy, lambda_Barcelona_grocery) +
box_cox(mob_parks, lambda_Barcelona_parks) +
box_cox(mob_residential, lambda_Barcelona_resd) +
box_cox(mob_retail_recreation, lambda_Barcelona_retail) +
box_cox(mob_transit_stations, lambda_Barcelona_transit) +
box_cox(mob_workplaces, lambda_Barcelona_work),
stepwise = FALSE, approx = FALSE),
Snaive = SNAIVE(box_cox(num_casos, lambda_Barcelona_casos))
)
# Show and report model
fit_model# A mable: 1 x 4
# Key: provincia [1]
provincia arima_at1
<chr> <model>
1 Barcelona <LM w/ ARIMA(1,0,2)(1,0,0)[7] errors>
# … with 2 more variables: arima_at2 <model>, Snaive <model>glance(fit_model)# A tibble: 3 × 9
provincia .model sigma2 log_lik AIC AICc BIC ar_roots ma_roots
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <list> <list>
1 Barcelona arima_at1 0.501 -525. 1076. 1077. 1131. <cpl [8]> <cpl [2]>
2 Barcelona arima_at2 0.492 -520. 1068. 1069. 1127. <cpl [9]> <cpl [2]>
3 Barcelona Snaive 3.23 NA NA NA NA <NULL> <NULL> fit_model %>%
pivot_longer(-provincia,
names_to = ".model",
values_to = "orders") %>%
left_join(glance(fit_model), by = c("provincia", ".model")) %>%
arrange(AICc) %>%
select(.model:BIC)# A mable: 3 x 7
# Key: .model [3]
.model orders sigma2 log_lik AIC AICc BIC
<chr> <model> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_… <LM w/ ARIMA(2,0,2)(1,0,0)[7] errors> 0.492 -520. 1068. 1069. 1127.
2 arima_… <LM w/ ARIMA(1,0,2)(1,0,0)[7] errors> 0.501 -525. 1076. 1077. 1131.
3 Snaive <SNAIVE> 3.23 NA NA NA NA # We use a Ljung-Box test >> large p-value, confirms residuals are similar / considered to white noise
fit_model %>%
select(Snaive) %>%
gg_tsresiduals()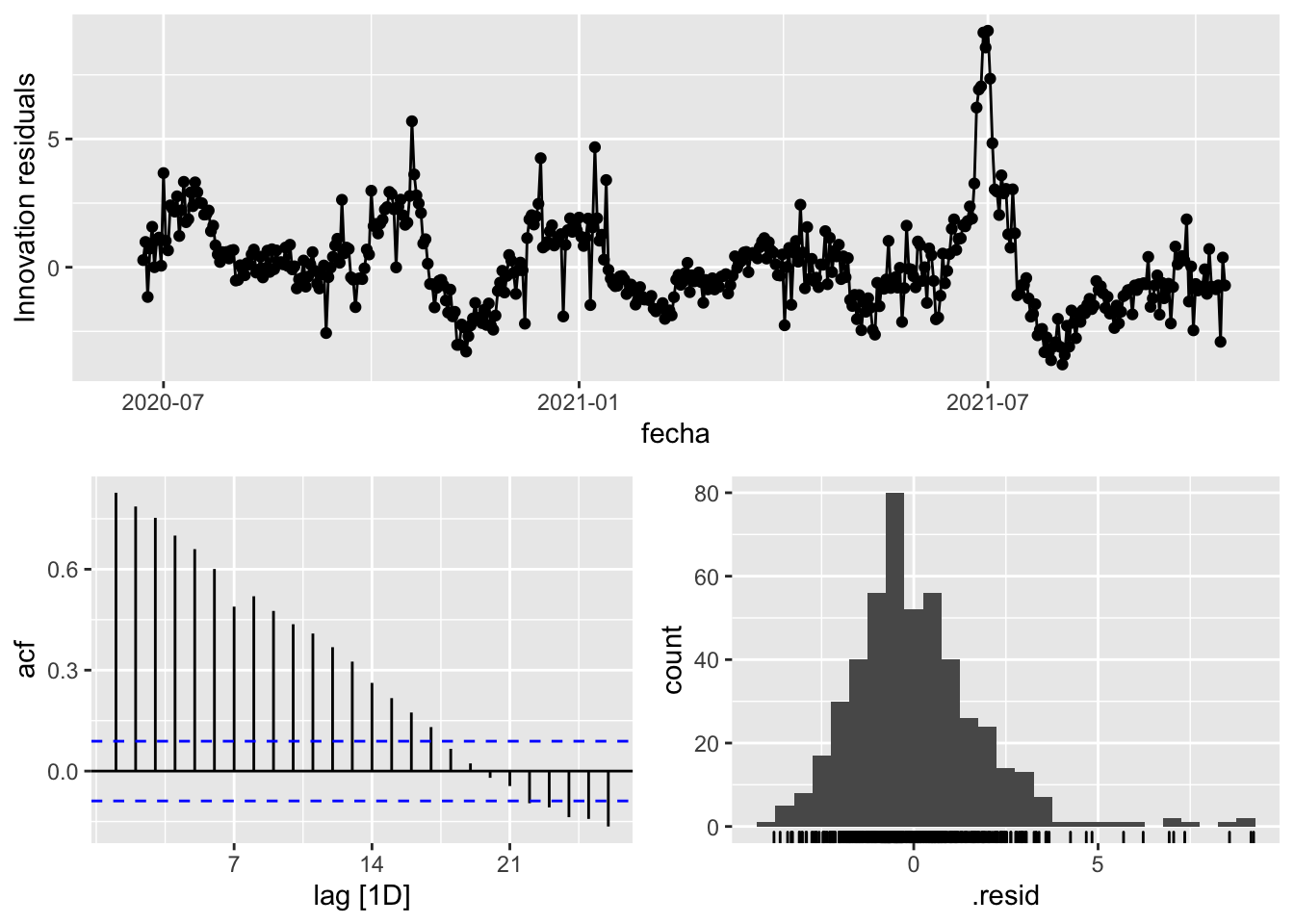
fit_model %>%
select(arima_at1) %>%
gg_tsresiduals()
fit_model %>%
select(arima_at2) %>%
gg_tsresiduals()
augment(fit_model) %>%
features(.innov, ljung_box, lag=7)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Barcelona arima_at1 49.8 0.0000000156
2 Barcelona arima_at2 52.9 0.00000000385
3 Barcelona Snaive 1648. 0 augment(fit_model) %>%
features(.innov, ljung_box, lag=14)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Barcelona arima_at1 68.6 0.00000000350
2 Barcelona arima_at2 69.2 0.00000000268
3 Barcelona Snaive 2222. 0 augment(fit_model) %>%
features(.innov, ljung_box, lag=21)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Barcelona arima_at1 77.6 0.0000000204
2 Barcelona arima_at2 78.4 0.0000000147
3 Barcelona Snaive 2272. 0 augment(fit_model) %>%
features(.innov, ljung_box, lag=90)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Barcelona arima_at1 182. 0.0000000314
2 Barcelona arima_at2 176. 0.000000148
3 Barcelona Snaive 3575. 0 # To predict future values of infections, we need to pass the model future/prediction values of the complementary variables tmed and mobility
# We are going to select those from the test dataset
data_fc_h7 <- data_Barcelona_test %>%
select(-num_casos) %>%
slice(1:7)
data_fc_h14 <- data_Barcelona_test %>%
select(-num_casos) %>%
slice(1:14)
data_fc_h21 <- data_Barcelona_test %>%
select(-num_casos) %>%
slice(1:21)
data_fc_h90 <- data_Barcelona_test %>%
select(-num_casos) %>%
slice(1:90)# Significant spikes out of 30 is still consistent with white noise
# To be sure, use a Ljung-Box test, which has a large p-value, confirming that
# the residuals are similar to white noise.
# Note that the alternative models also passed this test.
#
# Forecast
fc_h7 <- fabletools::forecast(fit_model, new_data = data_fc_h7)
fc_h14 <- fabletools::forecast(fit_model, new_data = data_fc_h14)
fc_h21 <- fabletools::forecast(fit_model, new_data = data_fc_h21)
fc_h90 <- fabletools::forecast(fit_model, new_data = data_fc_h90)7 days
# Plots
fc_h7 %>%
autoplot(
data_Barcelona_test %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(7))
) +
labs(y = "Nº cases", title = "Barcelona - forecast h7")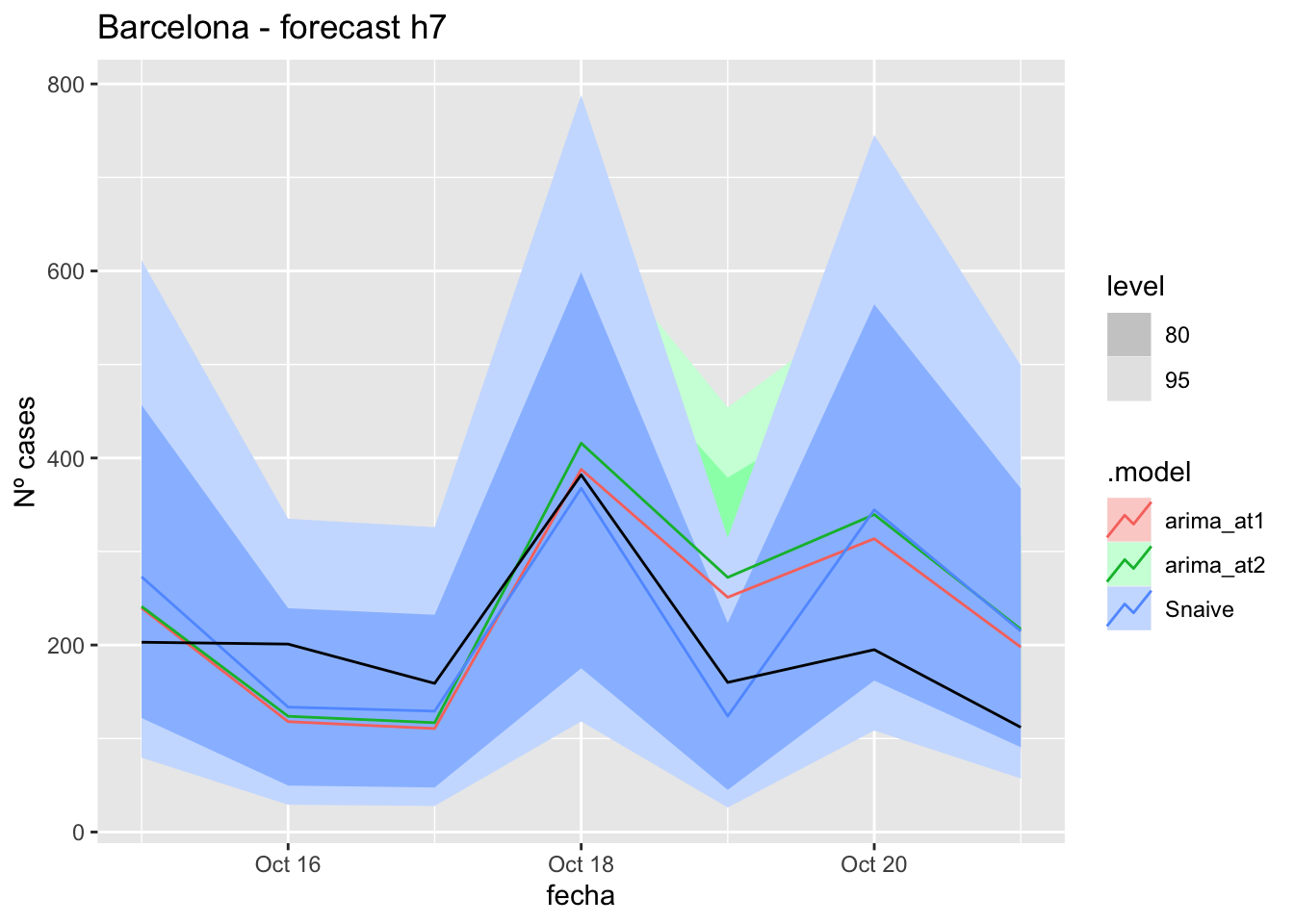
fc_h7 %>%
autoplot(
data_Barcelona %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(7) &
fecha > as.Date("2021-07-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Barcelona - forecast h7")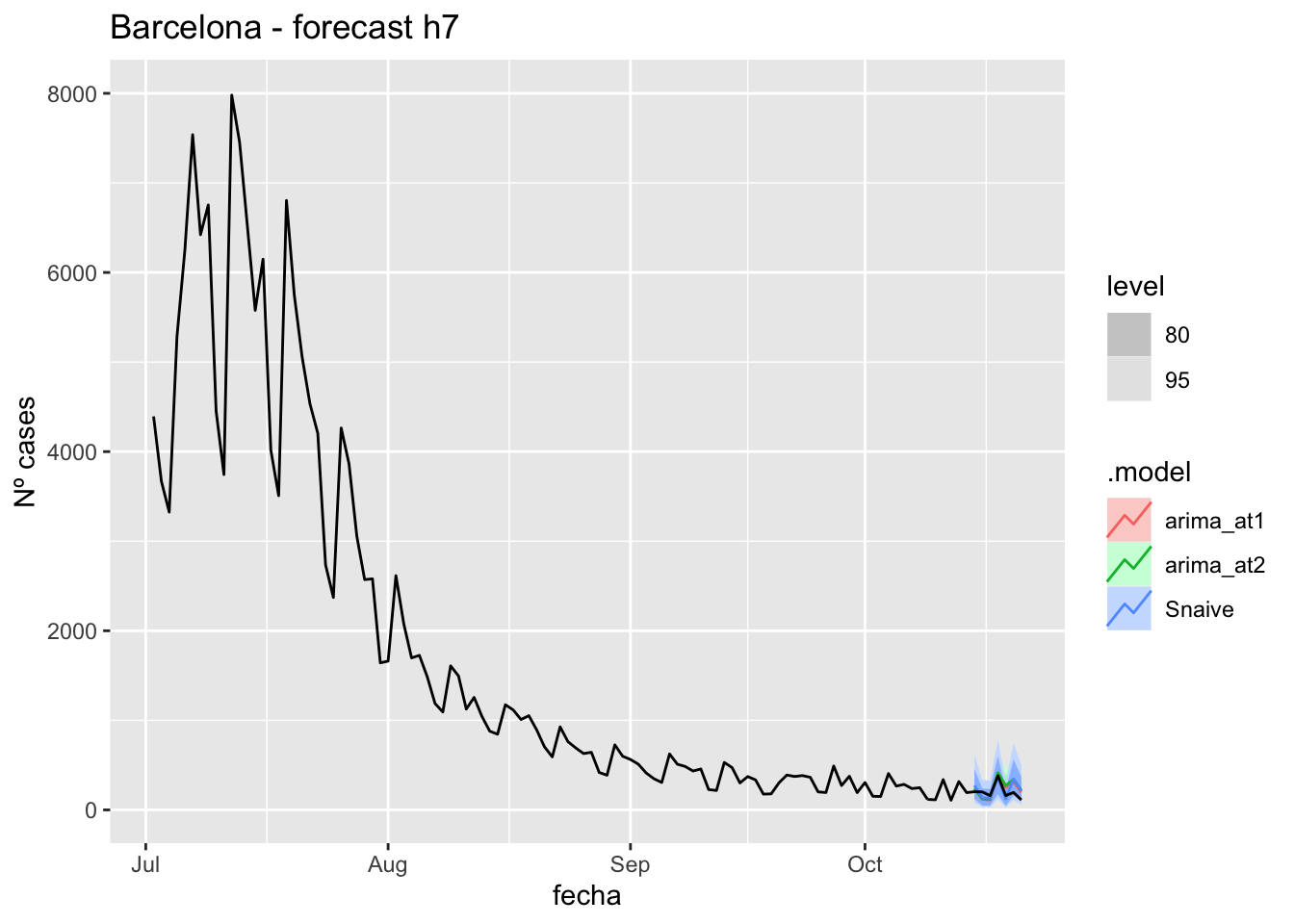
# Accuracy
fabletools::accuracy(fc_h7, data_Barcelona)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_at1 Barcelona Test -29.5 75.9 67.0 -20.3 40.8 0.178 0.114 0.551
2 arima_at2 Barcelona Test -45.0 88.5 79.0 -28.7 47.2 0.210 0.134 0.598
3 Snaive Barcelona Test -25.0 80.0 67.1 -17.8 40.2 0.178 0.121 0.18714 days
# Plots
fc_h14 %>%
autoplot(
data_Barcelona_test %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(14))
) +
labs(y = "Nº cases", title = "Barcelona - forecast h14")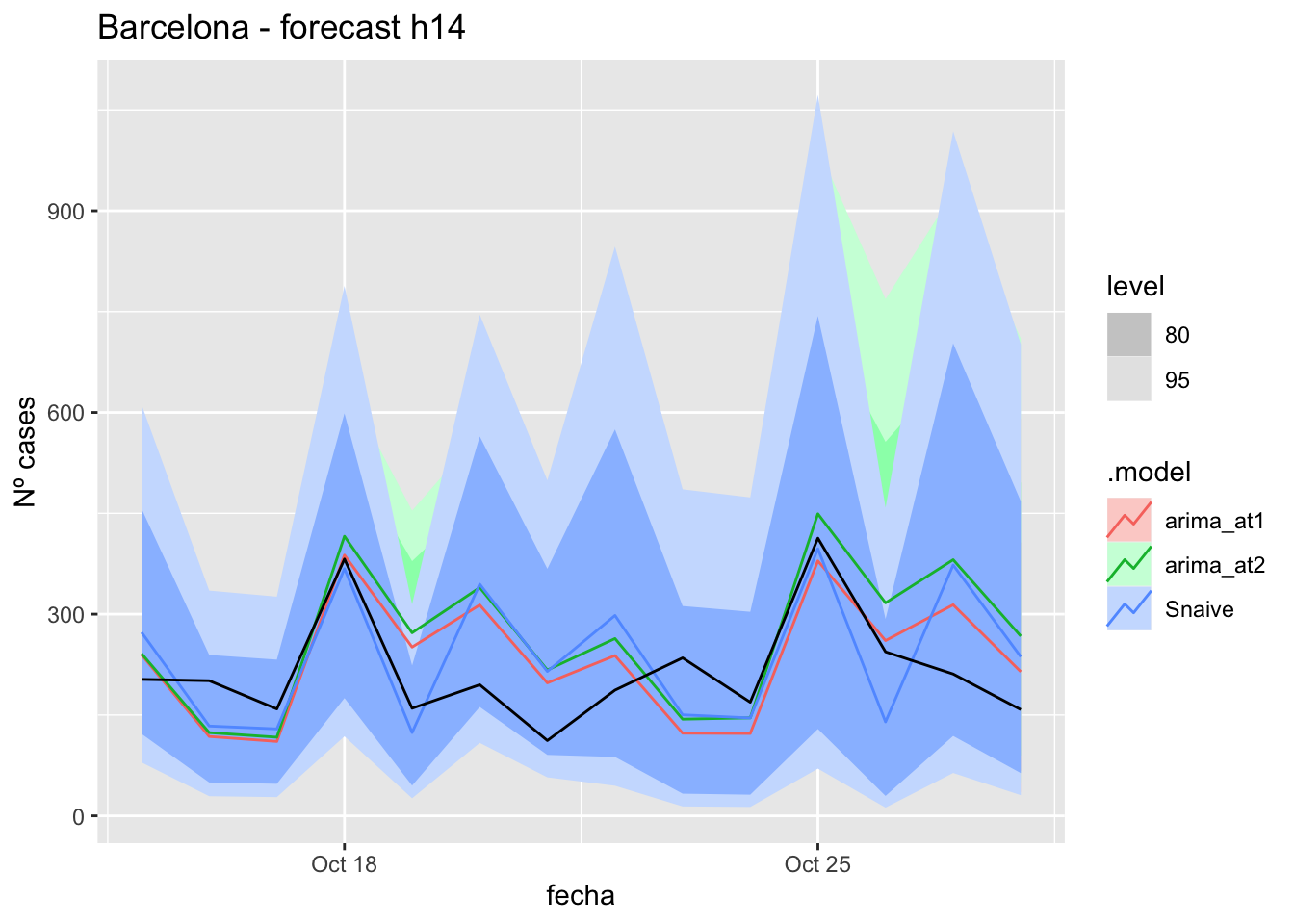
fc_h14 %>%
autoplot(
data_Barcelona %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(14) &
fecha > as.Date("2021-07-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Barcelona - forecast h14")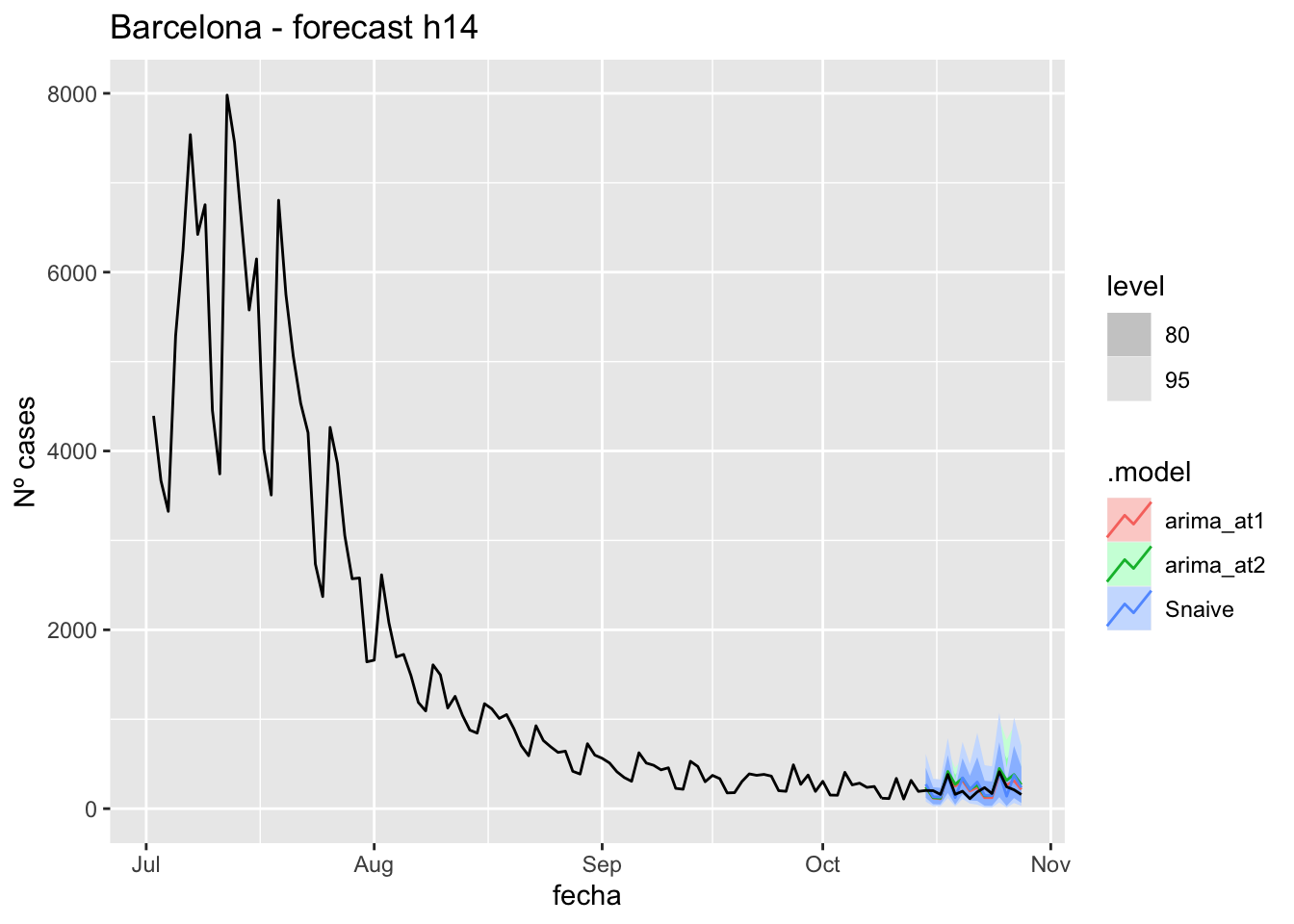
# Accuracy
fabletools::accuracy(fc_h14, data_Barcelona)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_at1 Barcelona Test -17.3 72.1 63.5 -12.7 34.8 0.169 0.109 0.462
2 arima_at2 Barcelona Test -47.5 91.4 80.9 -27.0 43.8 0.215 0.138 0.505
3 Snaive Barcelona Test -21.3 88.0 75.0 -15.3 40.3 0.199 0.133 0.063021 days
# Plots
fc_h21 %>%
autoplot(
data_Barcelona_test %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(21))
) +
labs(y = "Nº cases", title = "Barcelona - forecast h21")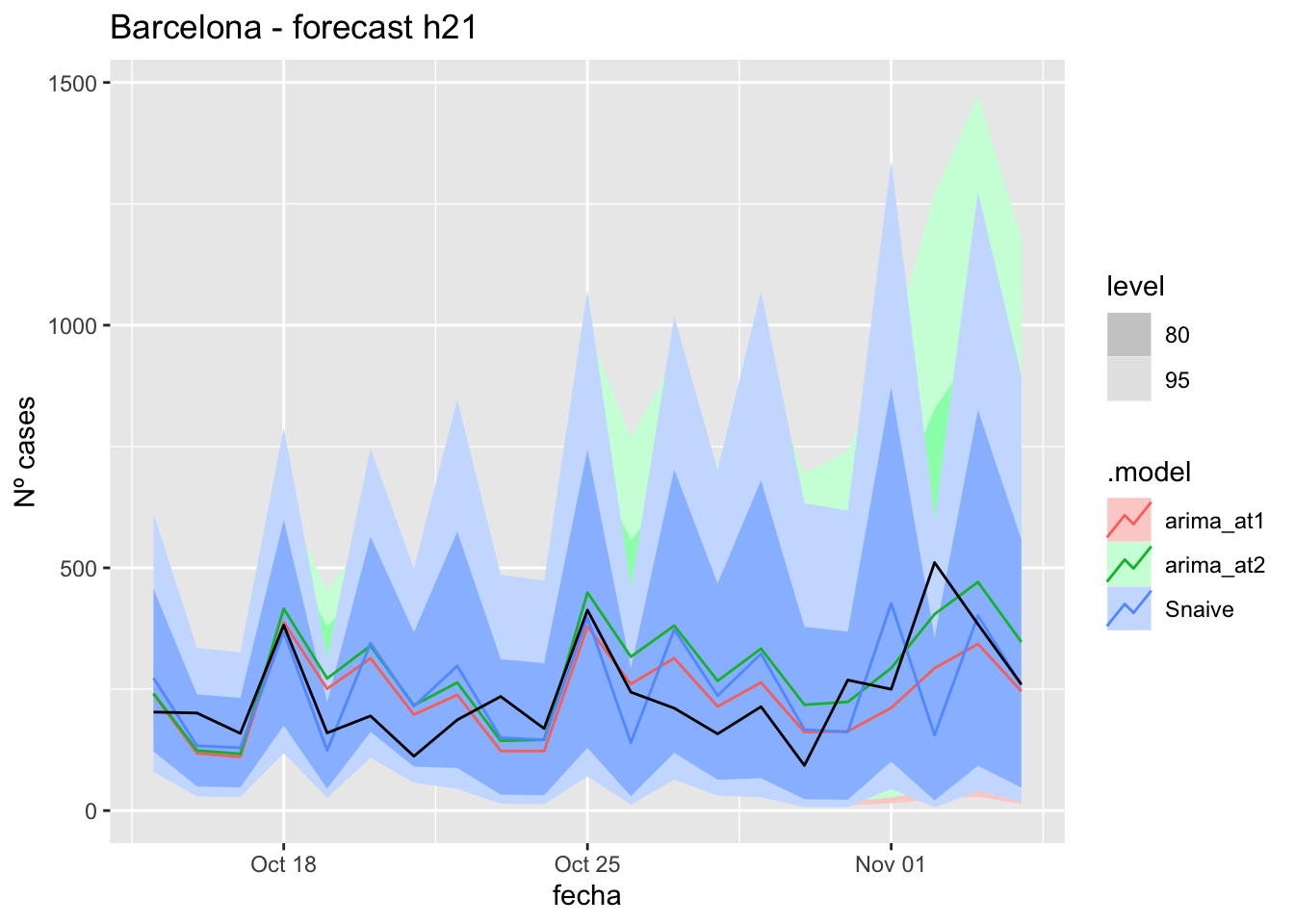
fc_h21 %>%
autoplot(
data_Barcelona %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(21) &
fecha > as.Date("2021-07-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Barcelona - forecast h21")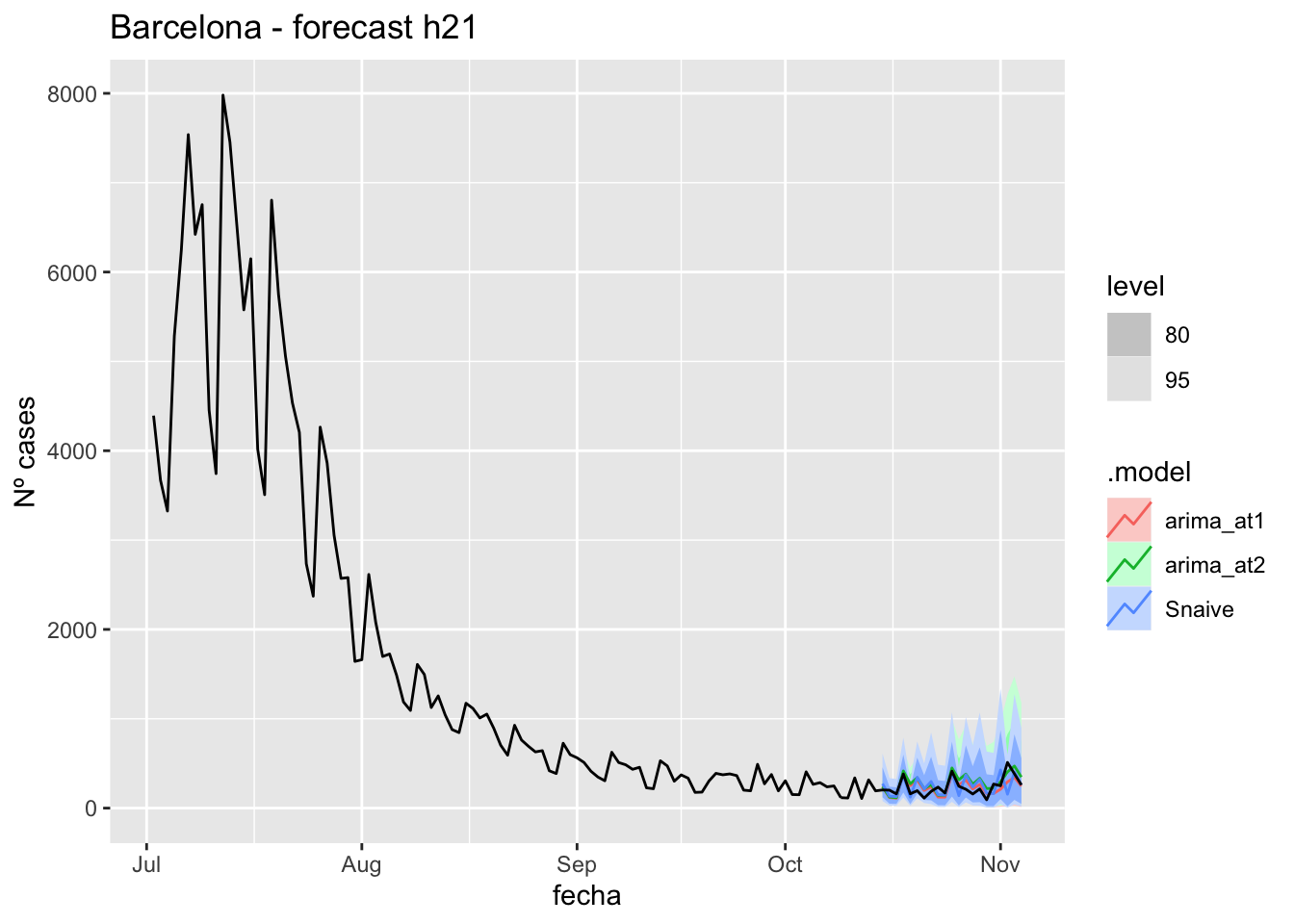
# Accuracy
fabletools::accuracy(fc_h21, data_Barcelona)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_at1 Barcelona Test 2.67 82.2 67.8 -7.70 33.2 0.180 0.124 0.389
2 arima_at2 Barcelona Test -46.5 91.9 83.2 -28.8 43.5 0.221 0.139 0.321
3 Snaive Barcelona Test -10.1 119. 90.1 -14.8 41.9 0.239 0.179 -0.24290 days
# Plots
fc_h90 %>%
autoplot(
data_Barcelona_test %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(90))
) +
labs(y = "Nº cases", title = "Barcelona - forecast h90")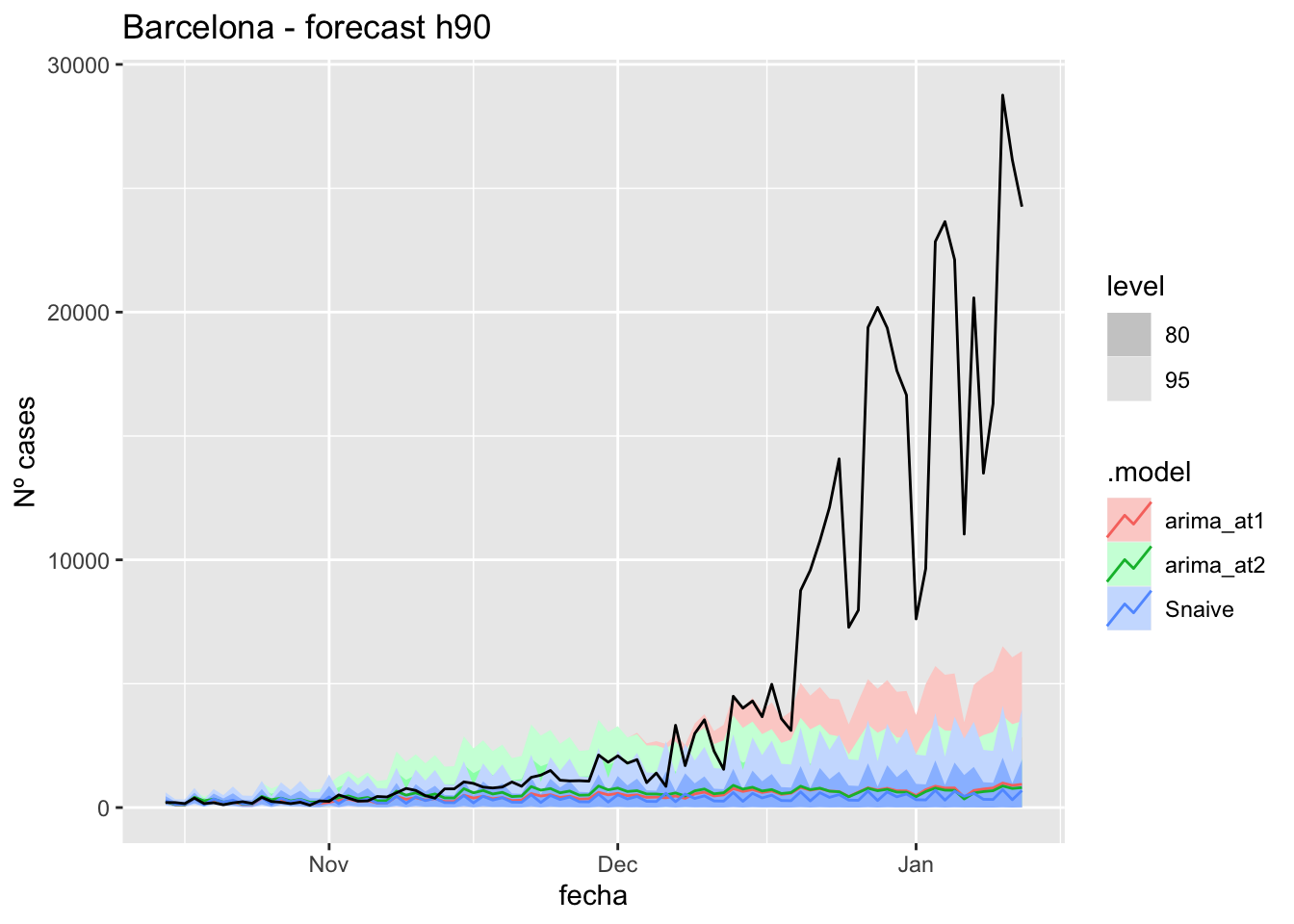
fc_h90 %>%
autoplot(
data_Barcelona %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(90) &
fecha > as.Date("2021-07-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Barcelona - forecast h90")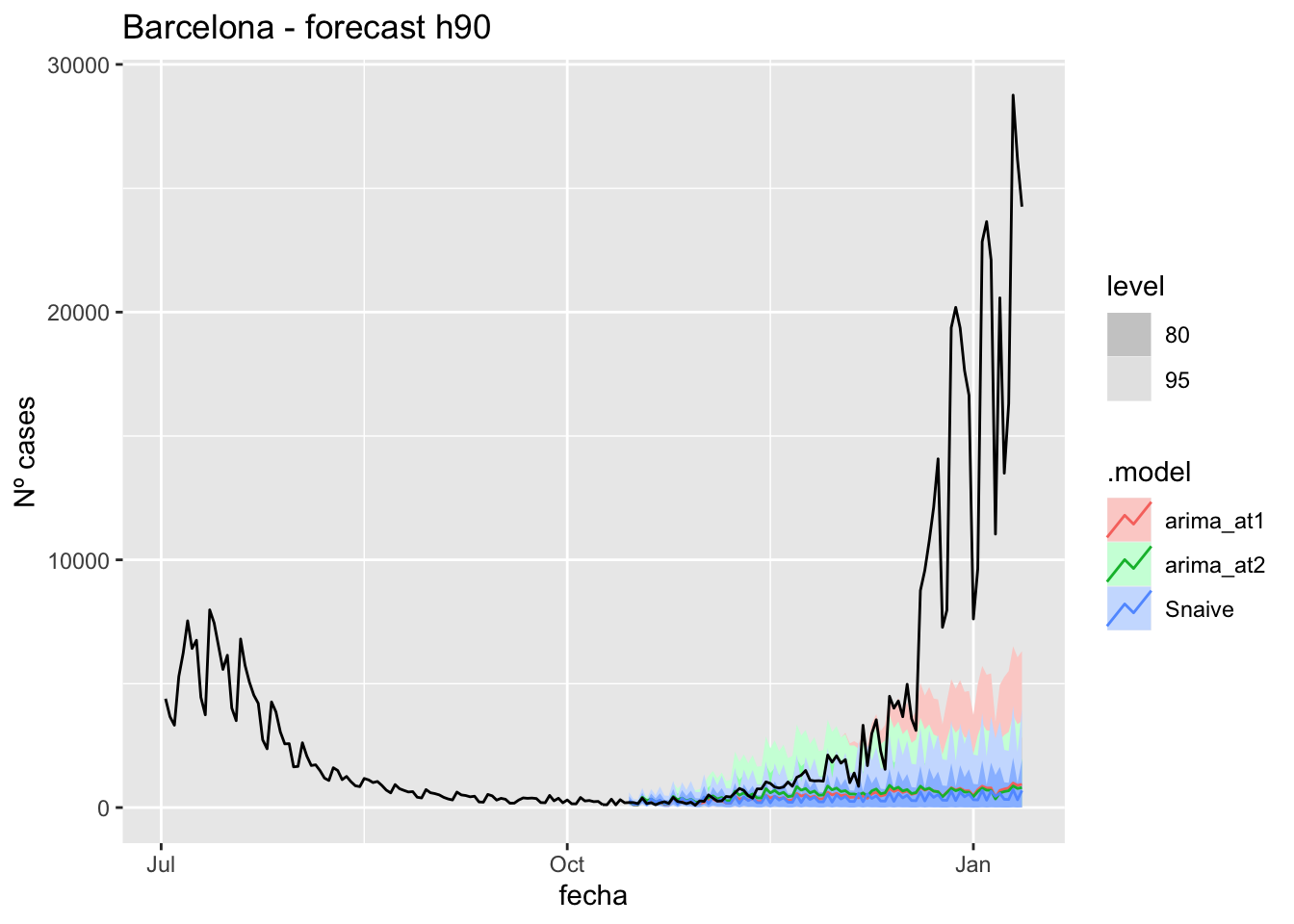
# Accuracy
fabletools::accuracy(fc_h90, data_Barcelona)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_at1 Barcelona Test 4772. 8726. 4788. 55.3 65.0 12.7 13.2 0.867
2 arima_at2 Barcelona Test 4698. 8750. 4736. 42.6 61.9 12.6 13.2 0.869
3 Snaive Barcelona Test 4888. 8865. 4913. 56.6 70.5 13.1 13.4 0.870Madrid
data_Madrid <- covid_data %>%
filter(provincia == "Madrid") %>%
filter(fecha > as.Date("2020-06-14", format = "%Y-%m-%d")) %>%
select(provincia, fecha, num_casos, tmed, mob_grocery_pharmacy, mob_parks, mob_residential, mob_retail_recreation, mob_transit_stations, mob_workplaces) %>%
drop_na() %>%
as_tsibble(key = provincia, index = fecha)
data_Madrid# A tsibble: 653 x 10 [1D]
# Key: provincia [1]
provincia fecha num_casos tmed mob_grocery_pharmacy mob_parks
<chr> <date> <dbl> <dbl> <dbl> <dbl>
1 Madrid 2020-06-15 153 20.7 -8 34
2 Madrid 2020-06-16 91 21 -8 21
3 Madrid 2020-06-17 93 20.2 -5 34
4 Madrid 2020-06-18 85 21.9 -7 30
5 Madrid 2020-06-19 78 22.6 -9 22
6 Madrid 2020-06-20 67 23.4 -12 25
7 Madrid 2020-06-21 42 25 -13 14
8 Madrid 2020-06-22 60 27.3 -8 29
9 Madrid 2020-06-23 68 28.4 -9 23
10 Madrid 2020-06-24 49 28 -7 22
# … with 643 more rows, and 4 more variables: mob_residential <dbl>,
# mob_retail_recreation <dbl>, mob_transit_stations <dbl>,
# mob_workplaces <dbl>data_Madrid_train <- data_Madrid %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d"))
summary(data_Madrid_train$fecha) Min. 1st Qu. Median Mean 3rd Qu. Max.
"2020-06-15" "2020-10-14" "2021-02-13" "2021-02-13" "2021-06-14" "2021-10-14" data_Madrid_test <- data_Madrid %>%
filter(fecha > as.Date("2021-10-14", format = "%Y-%m-%d"))
summary(data_Madrid_test$fecha) Min. 1st Qu. Median Mean 3rd Qu. Max.
"2021-10-15" "2021-11-25" "2022-01-05" "2022-01-05" "2022-02-15" "2022-03-29" # Lamba for num_casos
lambda_Madrid_casos <- data_asturias %>%
features(num_casos, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Madrid_casos[1] 0.2278229# Lamba for average temp
lambda_Madrid_tmed <- data_asturias %>%
features(tmed, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Madrid_tmed[1] 1.056074# Lamba for mobility
lambda_Madrid_grocery <- data_asturias %>%
features(mob_grocery_pharmacy, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Madrid_grocery[1] 1.377952lambda_Madrid_parks <- data_asturias %>%
features(mob_parks, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Madrid_parks[1] 0.7188469lambda_Madrid_resd <- data_asturias %>%
features(mob_residential, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Madrid_resd[1] 1.000064lambda_Madrid_retail <- data_asturias %>%
features(mob_retail_recreation, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Madrid_retail[1] 1.058286lambda_Madrid_transit <- data_asturias %>%
features(mob_transit_stations, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Madrid_transit[1] 1.116829lambda_Madrid_work <- data_asturias %>%
features(mob_workplaces, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Madrid_work[1] 1.999927fit_model <- data_Madrid_train %>%
model(
arima_at1 = ARIMA(box_cox(num_casos, lambda_Madrid_casos) ~
box_cox(tmed, lambda_Madrid_tmed) +
box_cox(mob_grocery_pharmacy, lambda_Madrid_grocery) +
box_cox(mob_parks, lambda_Madrid_parks) +
box_cox(mob_residential, lambda_Madrid_resd) +
box_cox(mob_retail_recreation, lambda_Madrid_retail) +
box_cox(mob_transit_stations, lambda_Madrid_transit) +
box_cox(mob_workplaces, lambda_Madrid_work)
),
arima_at2 = ARIMA(box_cox(num_casos, lambda_Madrid_casos) ~
box_cox(tmed, lambda_Madrid_tmed) +
box_cox(mob_grocery_pharmacy, lambda_Madrid_grocery) +
box_cox(mob_parks, lambda_Madrid_parks) +
box_cox(mob_residential, lambda_Madrid_resd) +
box_cox(mob_retail_recreation, lambda_Madrid_retail) +
box_cox(mob_transit_stations, lambda_Madrid_transit) +
box_cox(mob_workplaces, lambda_Madrid_work),
stepwise = FALSE, approx = FALSE),
Snaive = SNAIVE(box_cox(num_casos, lambda_Madrid_casos))
)
# Show and report model
fit_model# A mable: 1 x 4
# Key: provincia [1]
provincia arima_at1
<chr> <model>
1 Madrid <LM w/ ARIMA(3,0,0)(1,0,1)[7] errors>
# … with 2 more variables: arima_at2 <model>, Snaive <model>glance(fit_model)# A tibble: 3 × 9
provincia .model sigma2 log_lik AIC AICc BIC ar_roots ma_roots
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <list> <list>
1 Madrid arima_at1 0.494 -519. 1066. 1067. 1125. <cpl [10]> <cpl [7]>
2 Madrid arima_at2 0.494 -519. 1066. 1067. 1125. <cpl [10]> <cpl [7]>
3 Madrid Snaive 3.02 NA NA NA NA <NULL> <NULL> fit_model %>%
pivot_longer(-provincia,
names_to = ".model",
values_to = "orders") %>%
left_join(glance(fit_model), by = c("provincia", ".model")) %>%
arrange(AICc) %>%
select(.model:BIC)# A mable: 3 x 7
# Key: .model [3]
.model orders sigma2 log_lik AIC AICc BIC
<chr> <model> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_… <LM w/ ARIMA(3,0,0)(1,0,1)[7] errors> 0.494 -519. 1066. 1067. 1125.
2 arima_… <LM w/ ARIMA(3,0,0)(1,0,1)[7] errors> 0.494 -519. 1066. 1067. 1125.
3 Snaive <SNAIVE> 3.02 NA NA NA NA # We use a Ljung-Box test >> large p-value, confirms residuals are similar / considered to white noise
fit_model %>%
select(Snaive) %>%
gg_tsresiduals()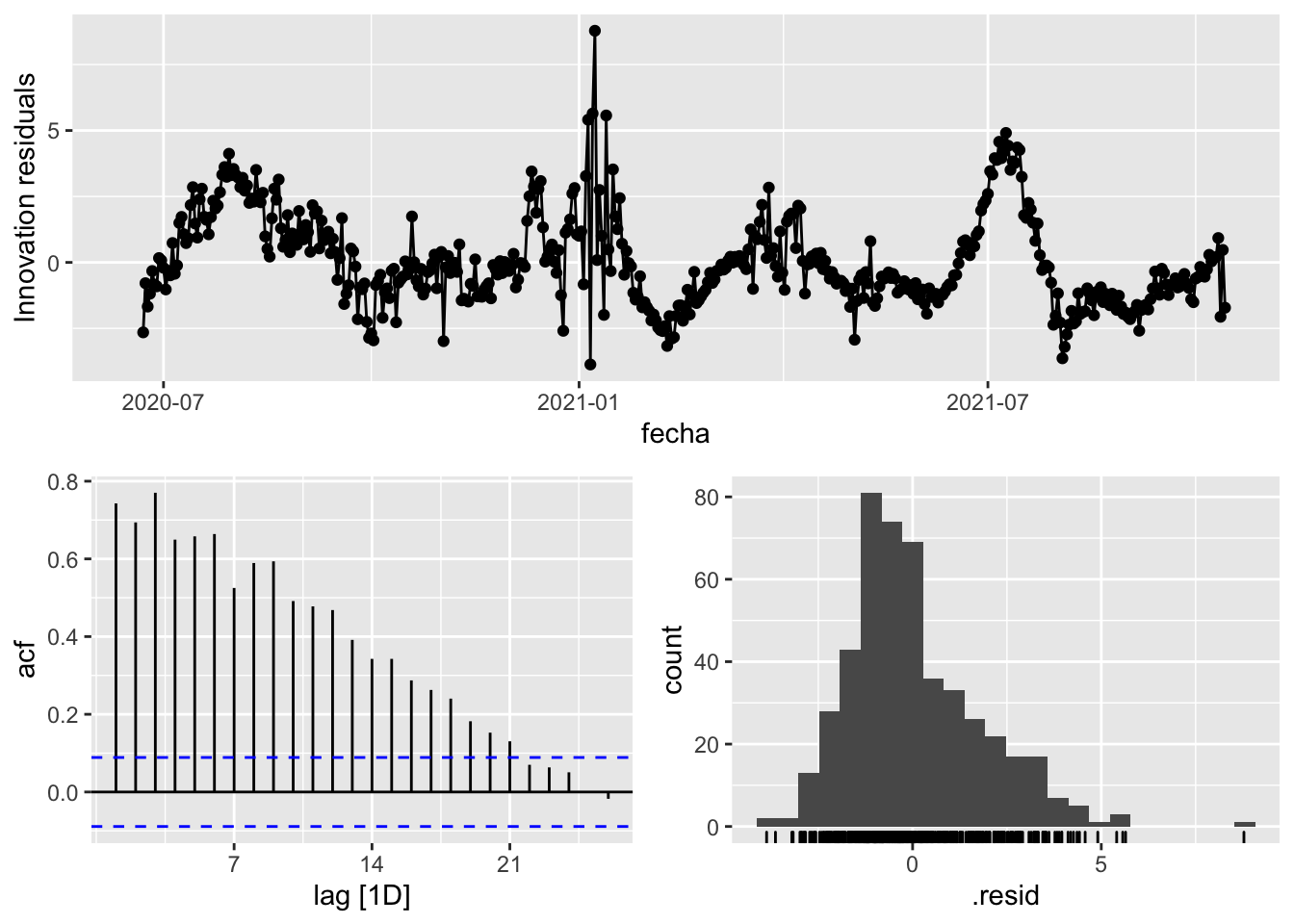
fit_model %>%
select(arima_at1) %>%
gg_tsresiduals()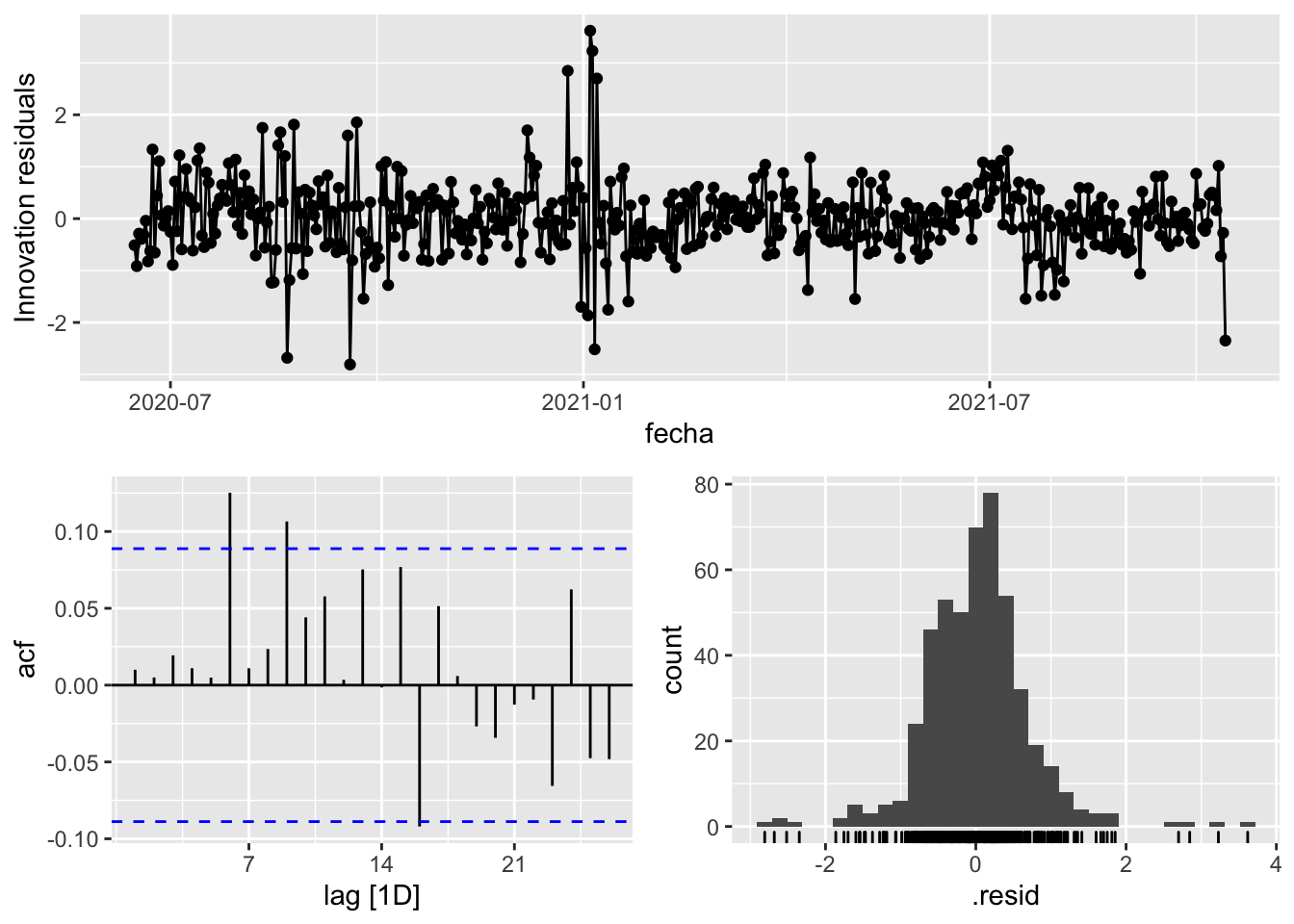
fit_model %>%
select(arima_at2) %>%
gg_tsresiduals()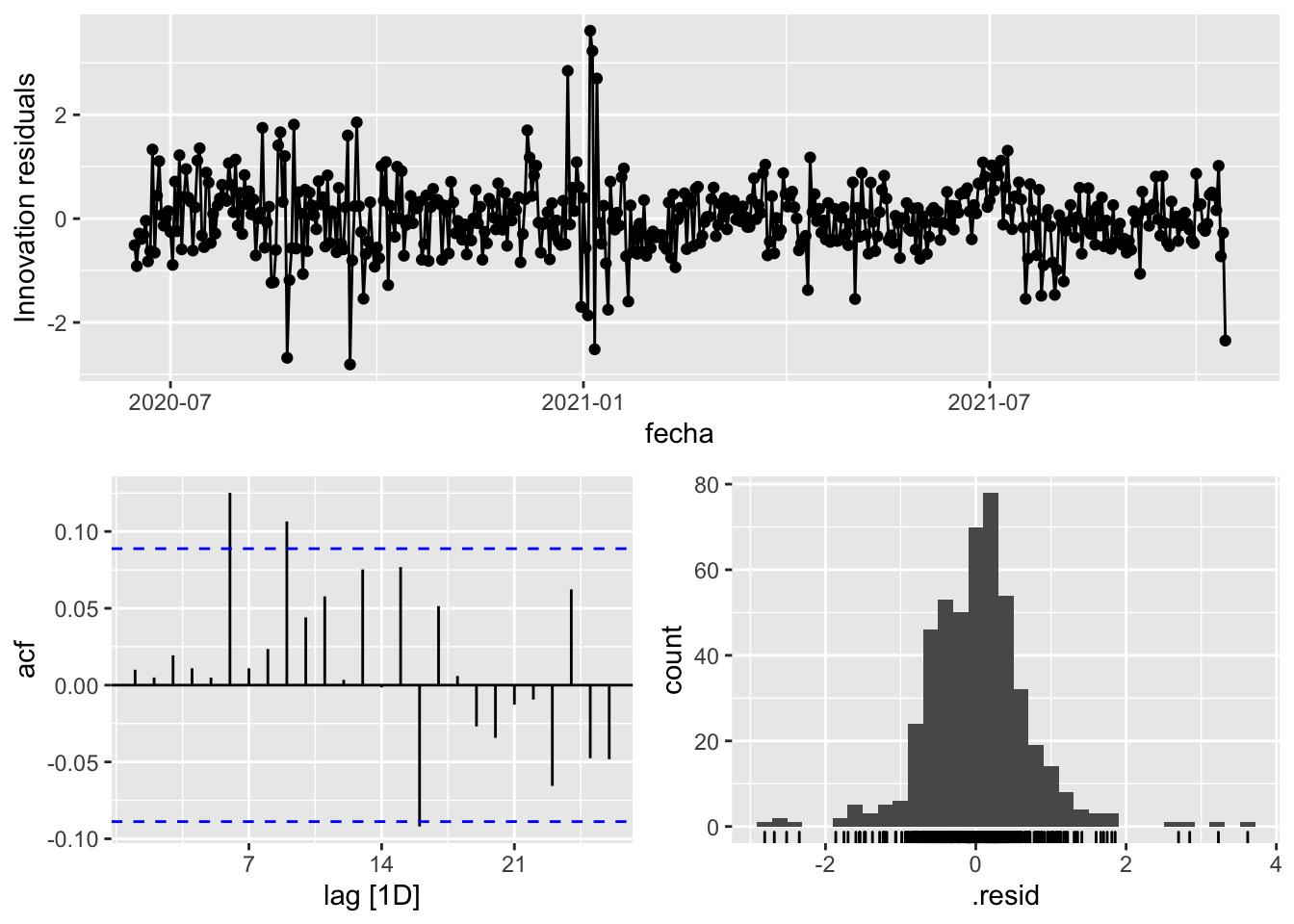
augment(fit_model) %>%
features(.innov, ljung_box, lag=7)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Madrid arima_at1 8.14 0.320
2 Madrid arima_at2 8.14 0.320
3 Madrid Snaive 1553. 0 augment(fit_model) %>%
features(.innov, ljung_box, lag=14)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Madrid arima_at1 19.6 0.144
2 Madrid arima_at2 19.6 0.144
3 Madrid Snaive 2371. 0 augment(fit_model) %>%
features(.innov, ljung_box, lag=21)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Madrid arima_at1 29.2 0.109
2 Madrid arima_at2 29.2 0.109
3 Madrid Snaive 2571. 0 augment(fit_model) %>%
features(.innov, ljung_box, lag=90)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Madrid arima_at1 119. 0.0225
2 Madrid arima_at2 119. 0.0225
3 Madrid Snaive 3699. 0 # To predict future values of infections, we need to pass the model future/prediction values of the complementary variables tmed and mobility
# We are going to select those from the test dataset
data_fc_h7 <- data_Madrid_test %>%
select(-num_casos) %>%
slice(1:7)
data_fc_h14 <- data_Madrid_test %>%
select(-num_casos) %>%
slice(1:14)
data_fc_h21 <- data_Madrid_test %>%
select(-num_casos) %>%
slice(1:21)
data_fc_h90 <- data_Madrid_test %>%
select(-num_casos) %>%
slice(1:90)# Significant spikes out of 30 is still consistent with white noise
# To be sure, use a Ljung-Box test, which has a large p-value, confirming that
# the residuals are similar to white noise.
# Note that the alternative models also passed this test.
#
# Forecast
fc_h7 <- fabletools::forecast(fit_model, new_data = data_fc_h7)
fc_h14 <- fabletools::forecast(fit_model, new_data = data_fc_h14)
fc_h21 <- fabletools::forecast(fit_model, new_data = data_fc_h21)
fc_h90 <- fabletools::forecast(fit_model, new_data = data_fc_h90)7 days
# Plots
fc_h7 %>%
autoplot(
data_Madrid_test %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(7))
) +
labs(y = "Nº cases", title = "Madrid - forecast h7")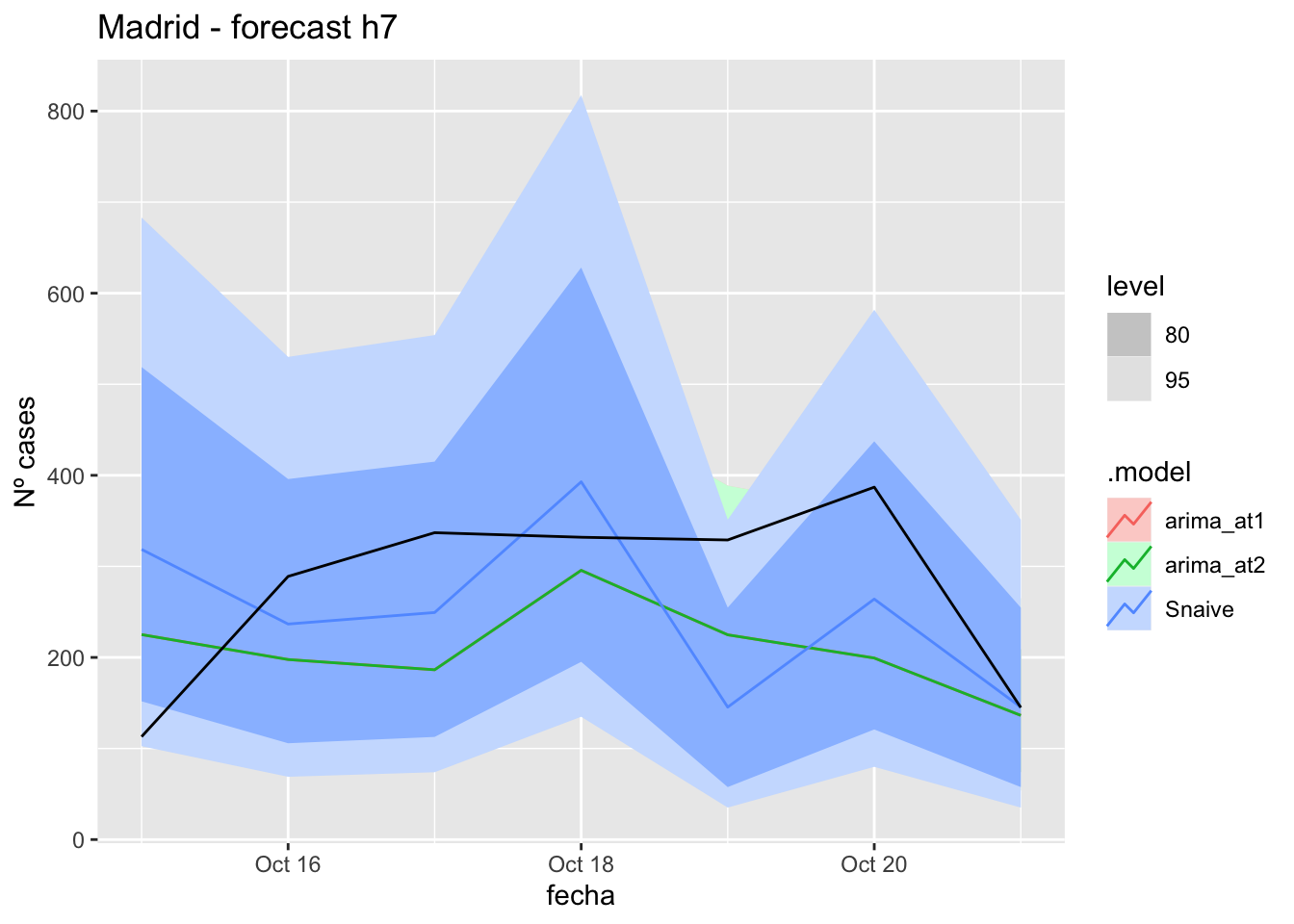
fc_h7 %>%
autoplot(
data_Madrid %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(7) &
fecha > as.Date("2021-07-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Madrid - forecast h7")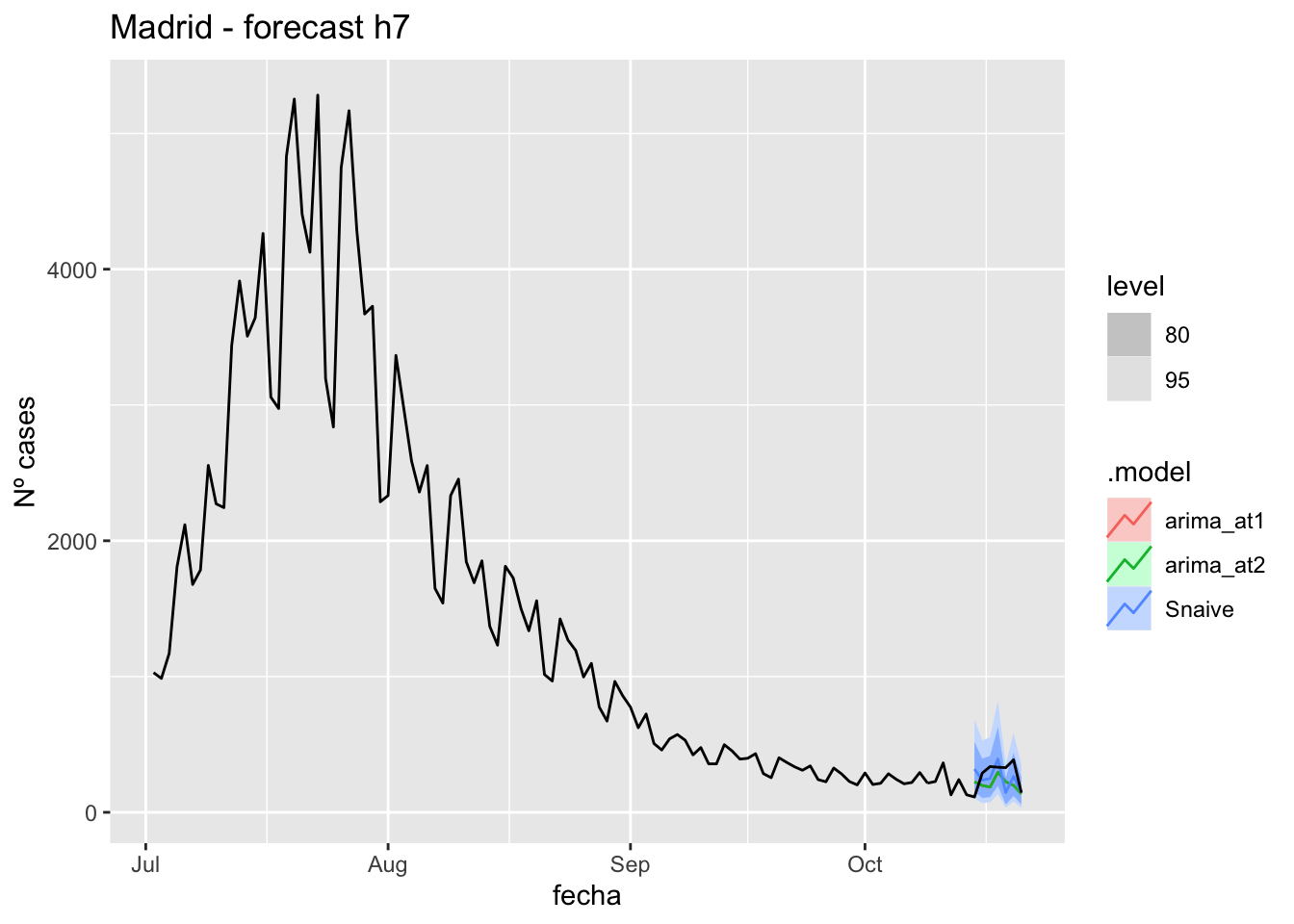
# Accuracy
fabletools::accuracy(fc_h7, data_Madrid)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_at1 Madrid Test 66.6 114. 98.7 10.6 38.9 0.233 0.170 -0.142
2 arima_at2 Madrid Test 66.6 114. 98.7 10.6 38.9 0.233 0.170 -0.142
3 Snaive Madrid Test 25.6 123. 102. -9.85 47.5 0.240 0.183 -0.10714 days
# Plots
fc_h14 %>%
autoplot(
data_Madrid_test %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(14))
) +
labs(y = "Nº cases", title = "Madrid - forecast h14")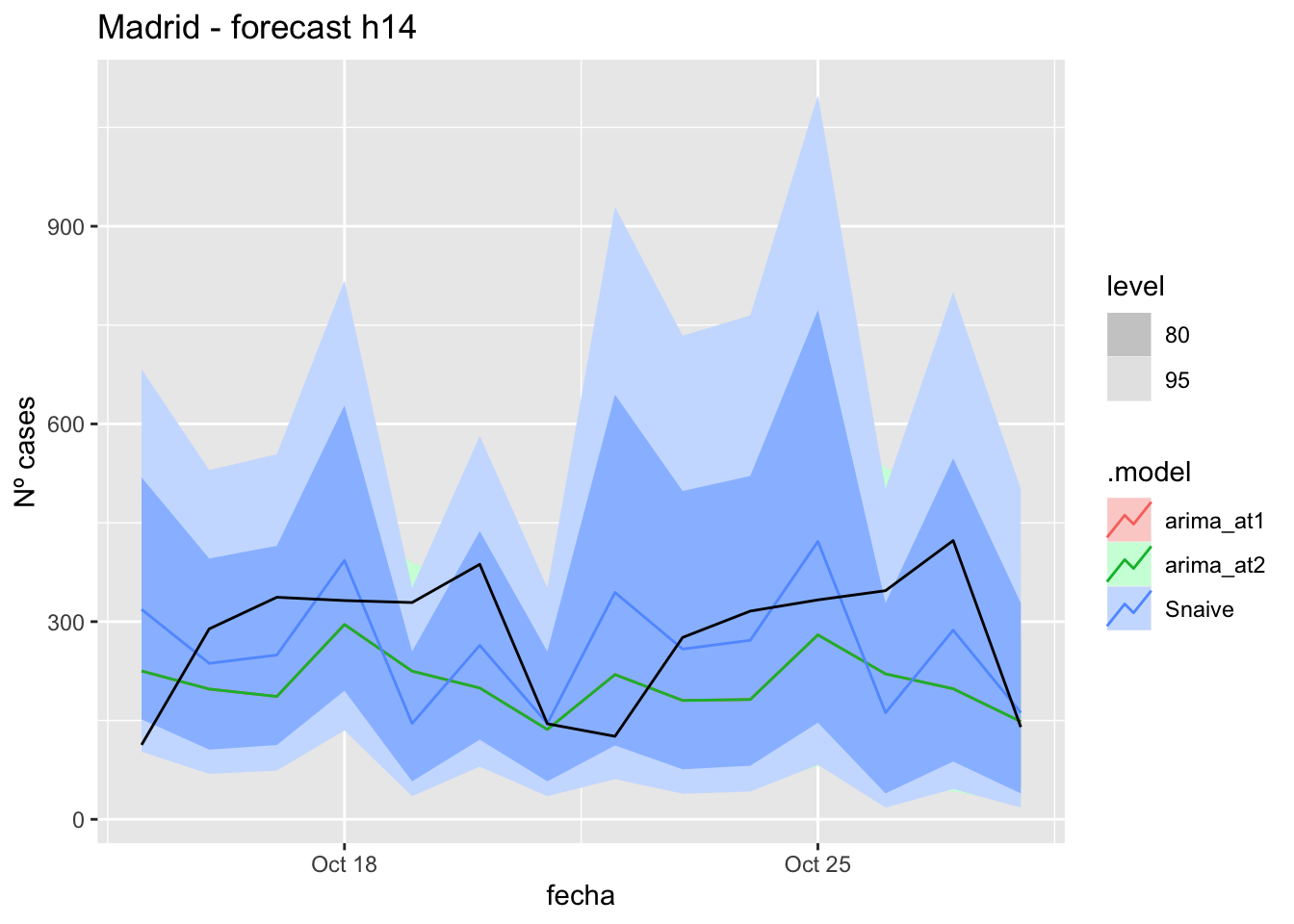
fc_h14 %>%
autoplot(
data_Madrid %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(14) &
fecha > as.Date("2021-07-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Madrid - forecast h14")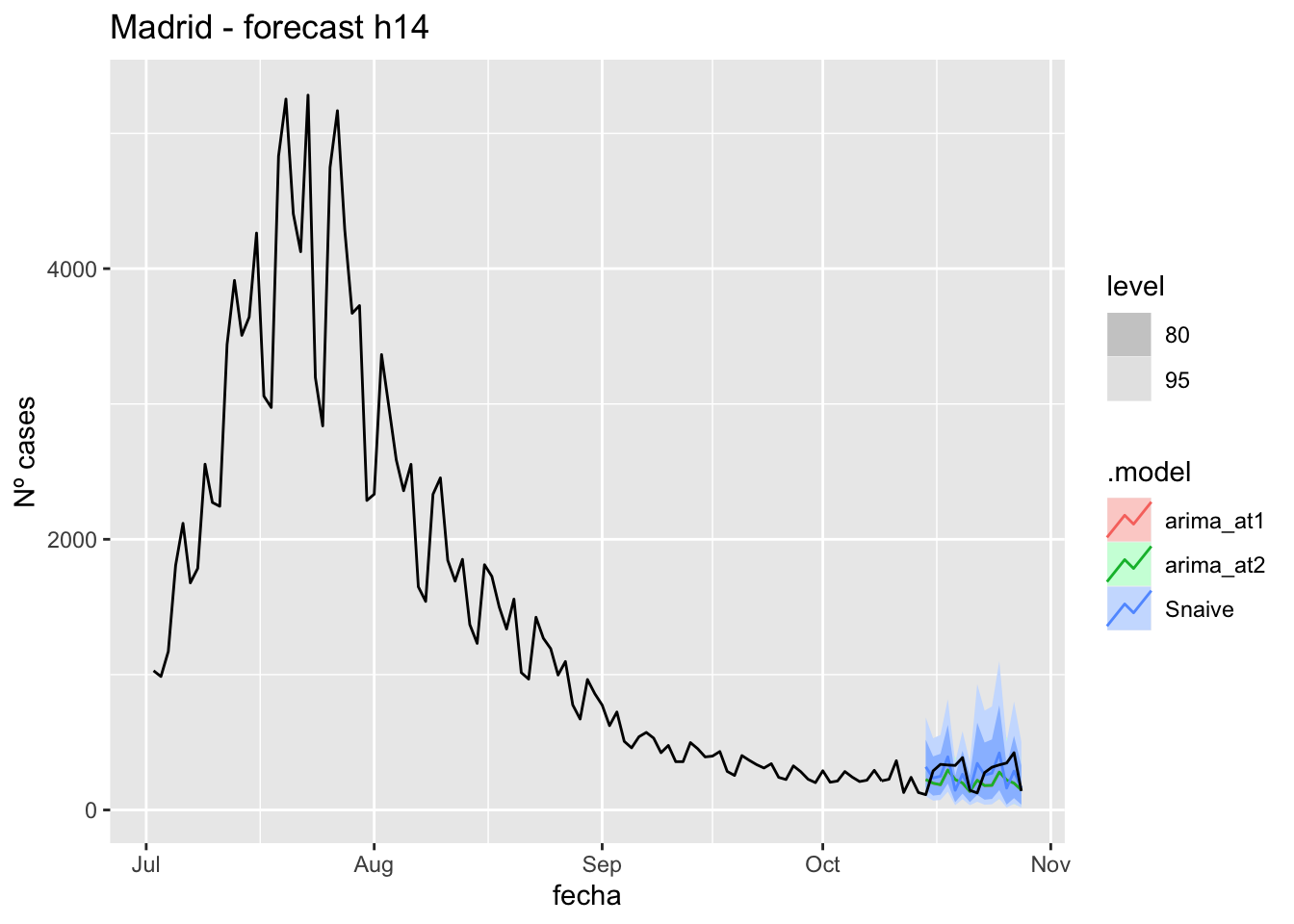
# Accuracy
fabletools::accuracy(fc_h14, data_Madrid)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_at1 Madrid Test 71.3 118. 102. 12.6 38.2 0.240 0.176 -0.0603
2 arima_at2 Madrid Test 71.3 118. 102. 12.6 38.2 0.240 0.176 -0.0603
3 Snaive Madrid Test 16.7 124. 102. -12.8 46.7 0.240 0.185 -0.043821 days
# Plots
fc_h21 %>%
autoplot(
data_Madrid_test %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(21))
) +
labs(y = "Nº cases", title = "Madrid - forecast h21")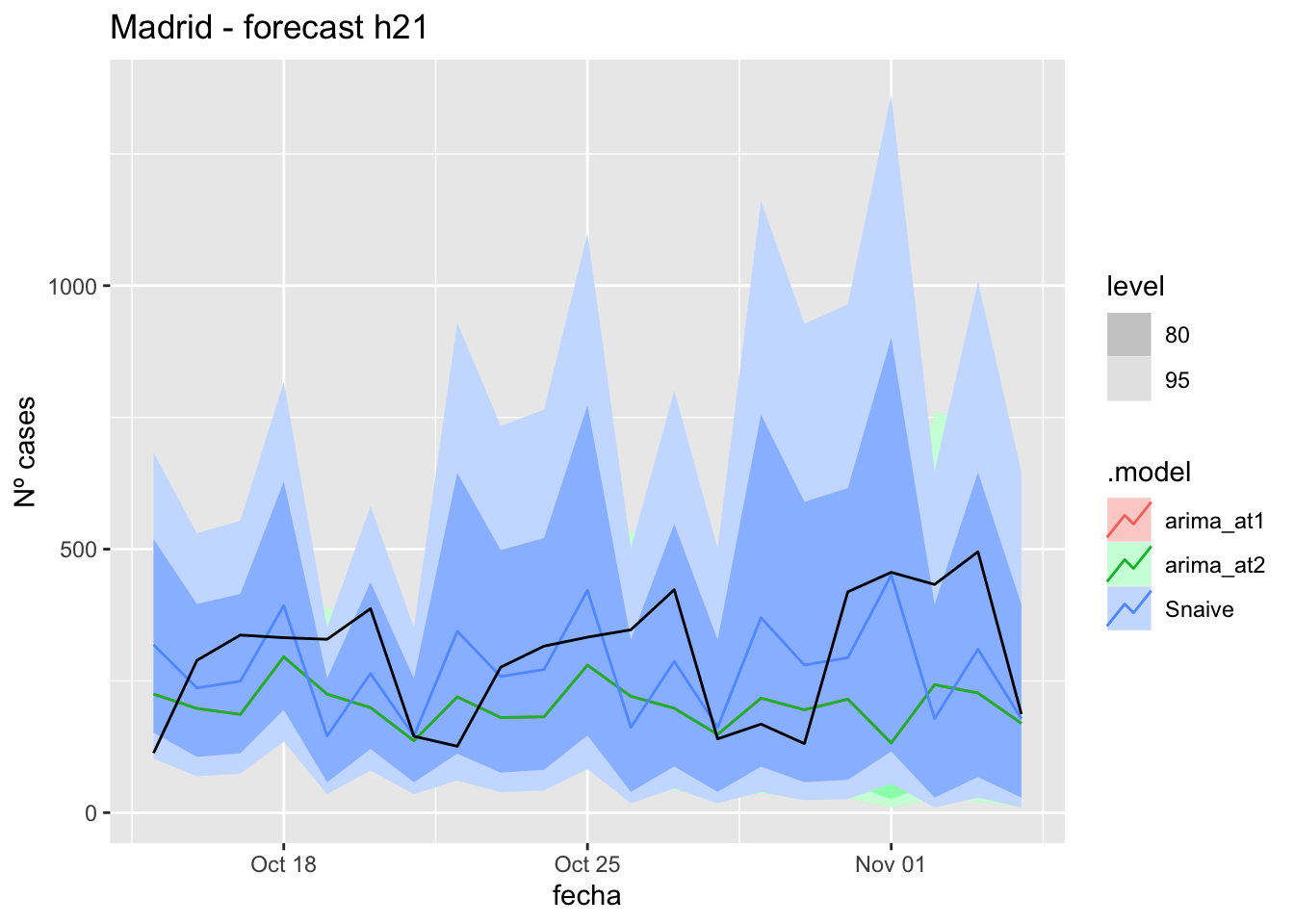
fc_h21 %>%
autoplot(
data_Madrid %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(21) &
fecha > as.Date("2021-07-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Madrid - forecast h21")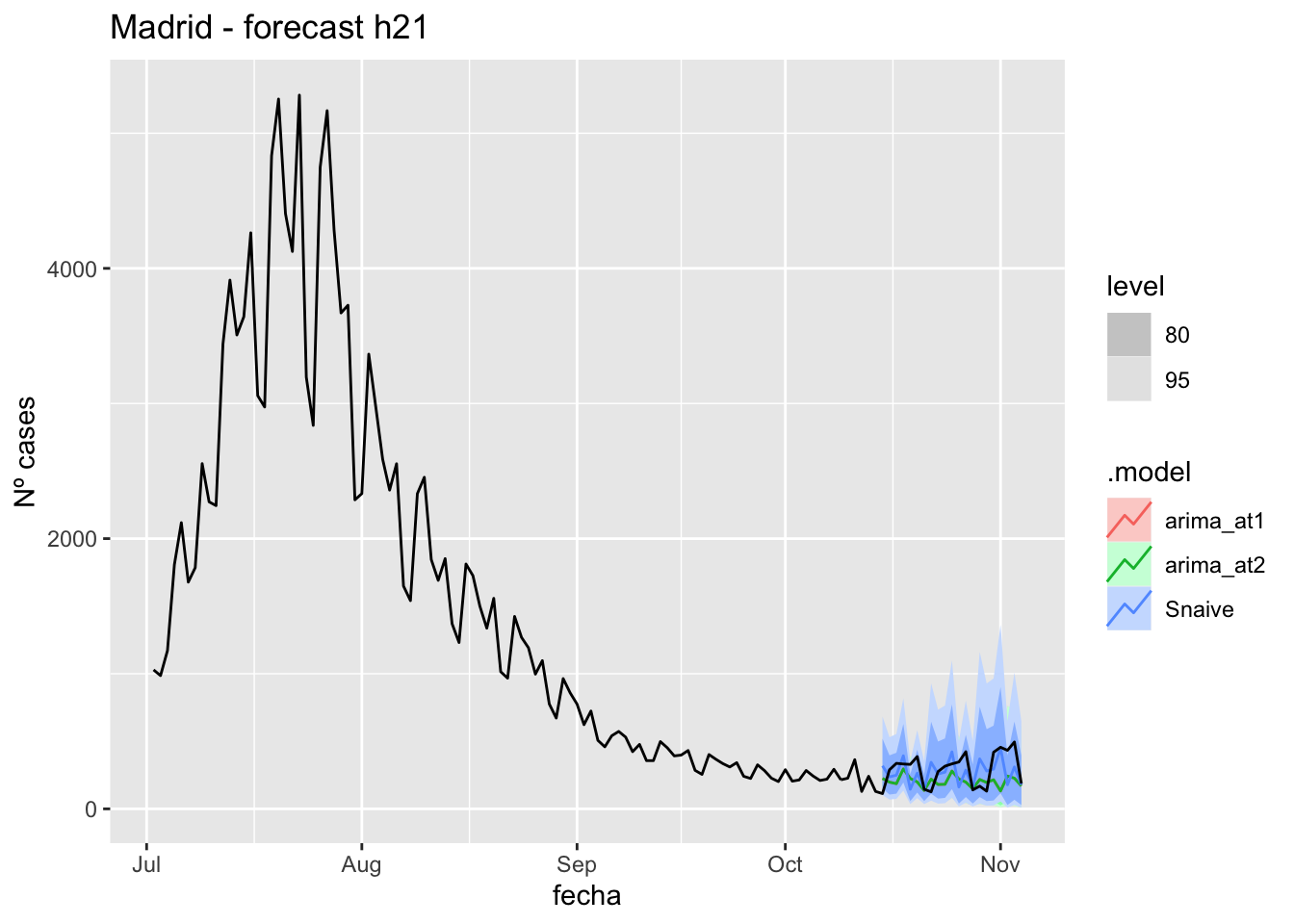
# Accuracy
fabletools::accuracy(fc_h21, data_Madrid)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_at1 Madrid Test 89.9 148. 121. 15.5 40.0 0.285 0.220 0.225
2 arima_at2 Madrid Test 89.9 148. 121. 15.5 40.0 0.285 0.220 0.225
3 Snaive Madrid Test 22.0 137. 112. -13.4 48.6 0.264 0.204 0.13190 days
# Plots
fc_h90 %>%
autoplot(
data_Madrid_test %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(90))
) +
labs(y = "Nº cases", title = "Madrid - forecast h90")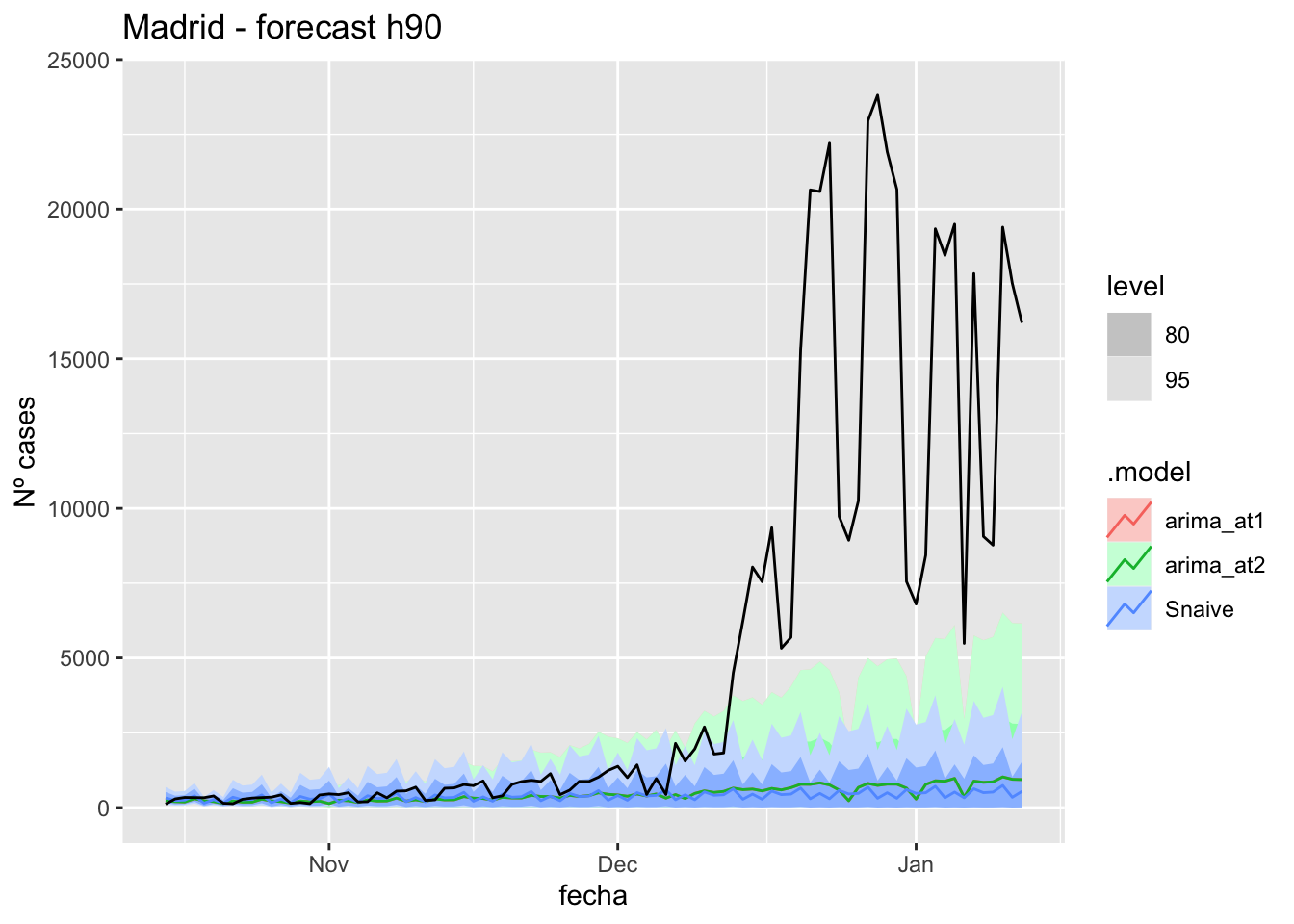
fc_h90 %>%
autoplot(
data_Madrid %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(90) &
fecha > as.Date("2021-07-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Madrid - forecast h90")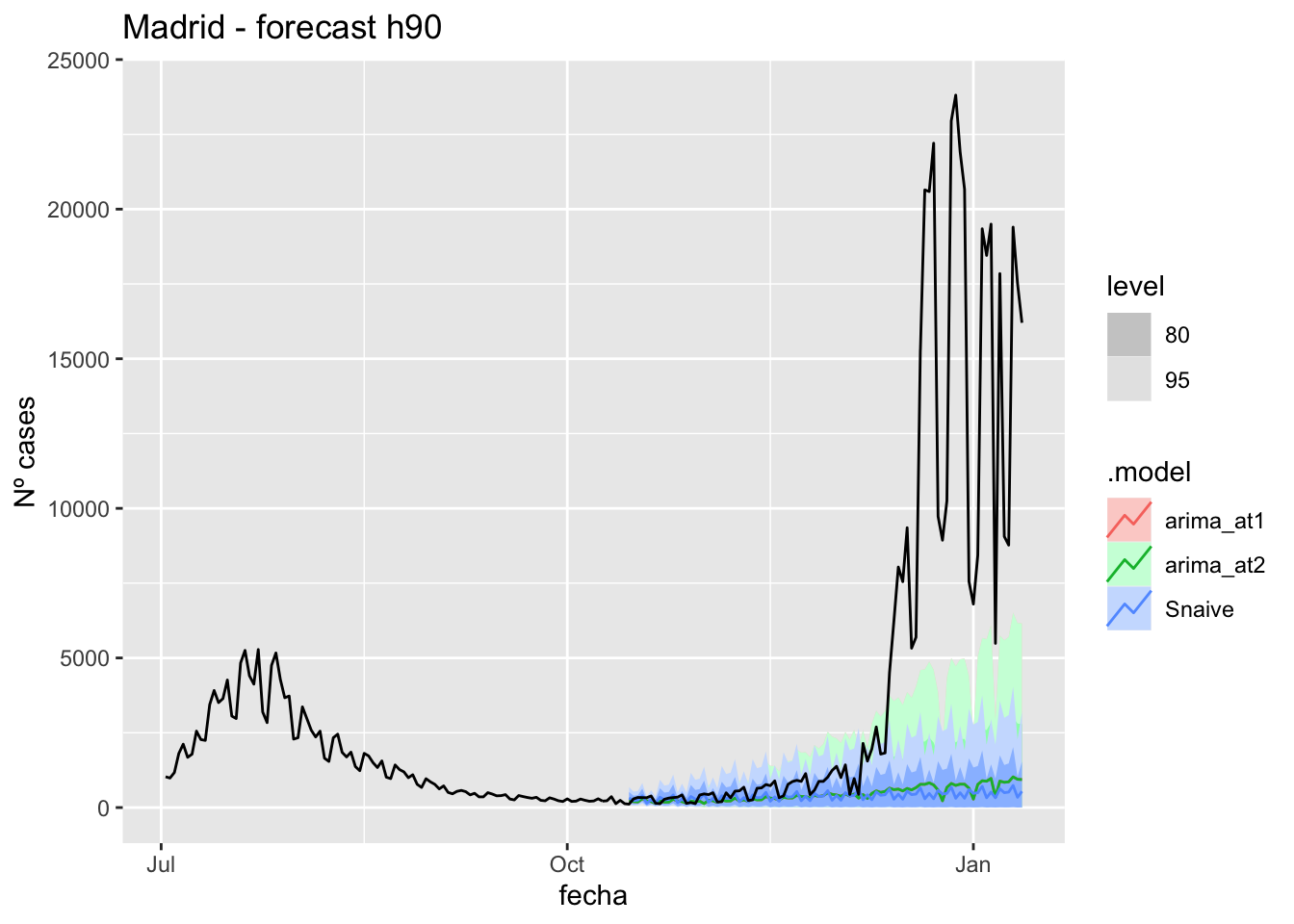
# Accuracy
fabletools::accuracy(fc_h90, data_Madrid)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_at1 Madrid Test 4656. 8366. 4665. 56.9 63.6 11.0 12.5 0.838
2 arima_at2 Madrid Test 4656. 8366. 4665. 56.9 63.6 11.0 12.5 0.838
3 Snaive Madrid Test 4718. 8525. 4752. 47.9 67.2 11.2 12.7 0.839Malaga
data_Malaga <- covid_data %>%
filter(provincia == "Málaga") %>%
filter(fecha > as.Date("2020-06-14", format = "%Y-%m-%d")) %>%
select(provincia, fecha, num_casos, tmed, mob_grocery_pharmacy, mob_parks, mob_residential, mob_retail_recreation, mob_transit_stations, mob_workplaces) %>%
drop_na() %>%
as_tsibble(key = provincia, index = fecha)
data_Malaga# A tsibble: 653 x 10 [1D]
# Key: provincia [1]
provincia fecha num_casos tmed mob_grocery_pharmacy mob_parks
<chr> <date> <dbl> <dbl> <dbl> <dbl>
1 Málaga 2020-06-15 1 27.2 -16 -9
2 Málaga 2020-06-16 1 27.8 -13 -4
3 Málaga 2020-06-17 2 26.9 -13 1
4 Málaga 2020-06-18 2 21.6 -12 3
5 Málaga 2020-06-19 1 24.7 -12 2
6 Málaga 2020-06-20 4 22.6 -14 8
7 Málaga 2020-06-21 4 22.7 -16 -3
8 Málaga 2020-06-22 6 25 -13 1
9 Málaga 2020-06-23 8 25.6 -10 18
10 Málaga 2020-06-24 65 24.5 -13 12
# … with 643 more rows, and 4 more variables: mob_residential <dbl>,
# mob_retail_recreation <dbl>, mob_transit_stations <dbl>,
# mob_workplaces <dbl>data_Malaga_train <- data_Malaga %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d"))
summary(data_Malaga_train$fecha) Min. 1st Qu. Median Mean 3rd Qu. Max.
"2020-06-15" "2020-10-14" "2021-02-13" "2021-02-13" "2021-06-14" "2021-10-14" data_Malaga_test <- data_Malaga %>%
filter(fecha > as.Date("2021-10-14", format = "%Y-%m-%d"))
summary(data_Malaga_test$fecha) Min. 1st Qu. Median Mean 3rd Qu. Max.
"2021-10-15" "2021-11-25" "2022-01-05" "2022-01-05" "2022-02-15" "2022-03-29" # Lamba for num_casos
lambda_Malaga_casos <- data_asturias %>%
features(num_casos, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Malaga_casos[1] 0.2278229# Lamba for average temp
lambda_Malaga_tmed <- data_asturias %>%
features(tmed, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Malaga_tmed[1] 1.056074# Lamba for mobility
lambda_Malaga_grocery <- data_asturias %>%
features(mob_grocery_pharmacy, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Malaga_grocery[1] 1.377952lambda_Malaga_parks <- data_asturias %>%
features(mob_parks, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Malaga_parks[1] 0.7188469lambda_Malaga_resd <- data_asturias %>%
features(mob_residential, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Malaga_resd[1] 1.000064lambda_Malaga_retail <- data_asturias %>%
features(mob_retail_recreation, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Malaga_retail[1] 1.058286lambda_Malaga_transit <- data_asturias %>%
features(mob_transit_stations, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Malaga_transit[1] 1.116829lambda_Malaga_work <- data_asturias %>%
features(mob_workplaces, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Malaga_work[1] 1.999927fit_model <- data_Malaga_train %>%
model(
arima_at1 = ARIMA(box_cox(num_casos, lambda_Malaga_casos) ~
box_cox(tmed, lambda_Malaga_tmed) +
box_cox(mob_grocery_pharmacy, lambda_Malaga_grocery) +
box_cox(mob_parks, lambda_Malaga_parks) +
box_cox(mob_residential, lambda_Malaga_resd) +
box_cox(mob_retail_recreation, lambda_Malaga_retail) +
box_cox(mob_transit_stations, lambda_Malaga_transit) +
box_cox(mob_workplaces, lambda_Malaga_work)
),
arima_at2 = ARIMA(box_cox(num_casos, lambda_Malaga_casos) ~
box_cox(tmed, lambda_Malaga_tmed) +
box_cox(mob_grocery_pharmacy, lambda_Malaga_grocery) +
box_cox(mob_parks, lambda_Malaga_parks) +
box_cox(mob_residential, lambda_Malaga_resd) +
box_cox(mob_retail_recreation, lambda_Malaga_retail) +
box_cox(mob_transit_stations, lambda_Malaga_transit) +
box_cox(mob_workplaces, lambda_Malaga_work),
stepwise = FALSE, approx = FALSE),
Snaive = SNAIVE(box_cox(num_casos, lambda_Malaga_casos))
)
# Show and report model
fit_model# A mable: 1 x 4
# Key: provincia [1]
provincia arima_at1
<chr> <model>
1 Málaga <LM w/ ARIMA(4,1,0)(2,0,0)[7] errors>
# … with 2 more variables: arima_at2 <model>, Snaive <model>glance(fit_model)# A tibble: 3 × 9
provincia .model sigma2 log_lik AIC AICc BIC ar_roots ma_roots
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <list> <list>
1 Málaga arima_at1 0.469 -500. 1029. 1030. 1087. <cpl [18]> <cpl [0]>
2 Málaga arima_at2 0.464 -498. 1024. 1025. 1083. <cpl [16]> <cpl [2]>
3 Málaga Snaive 2.08 NA NA NA NA <NULL> <NULL> fit_model %>%
pivot_longer(-provincia,
names_to = ".model",
values_to = "orders") %>%
left_join(glance(fit_model), by = c("provincia", ".model")) %>%
arrange(AICc) %>%
select(.model:BIC)# A mable: 3 x 7
# Key: .model [3]
.model orders sigma2 log_lik AIC AICc BIC
<chr> <model> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_… <LM w/ ARIMA(2,1,2)(2,0,0)[7] errors> 0.464 -498. 1024. 1025. 1083.
2 arima_… <LM w/ ARIMA(4,1,0)(2,0,0)[7] errors> 0.469 -500. 1029. 1030. 1087.
3 Snaive <SNAIVE> 2.08 NA NA NA NA # We use a Ljung-Box test >> large p-value, confirms residuals are similar / considered to white noise
fit_model %>%
select(Snaive) %>%
gg_tsresiduals()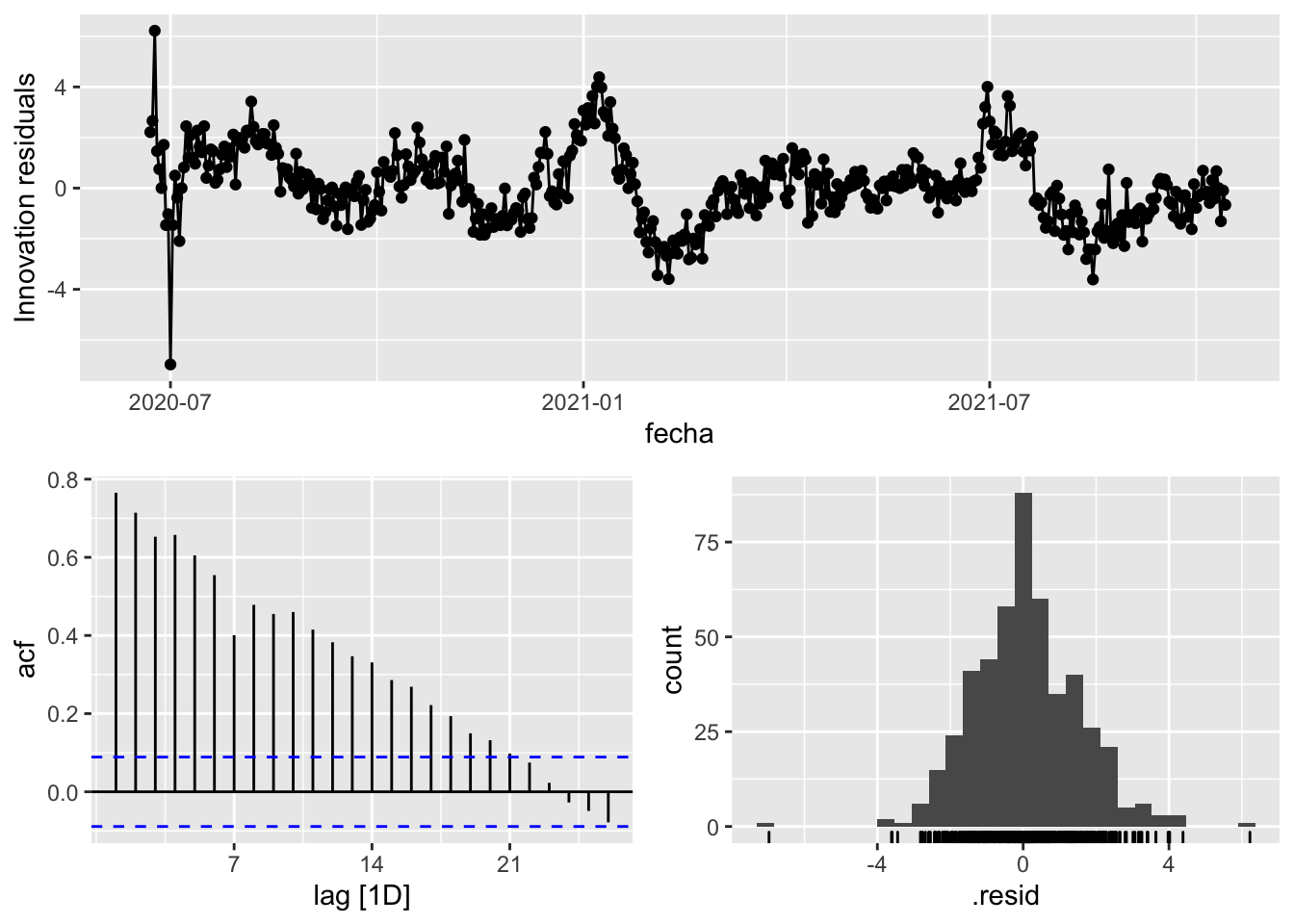
fit_model %>%
select(arima_at1) %>%
gg_tsresiduals()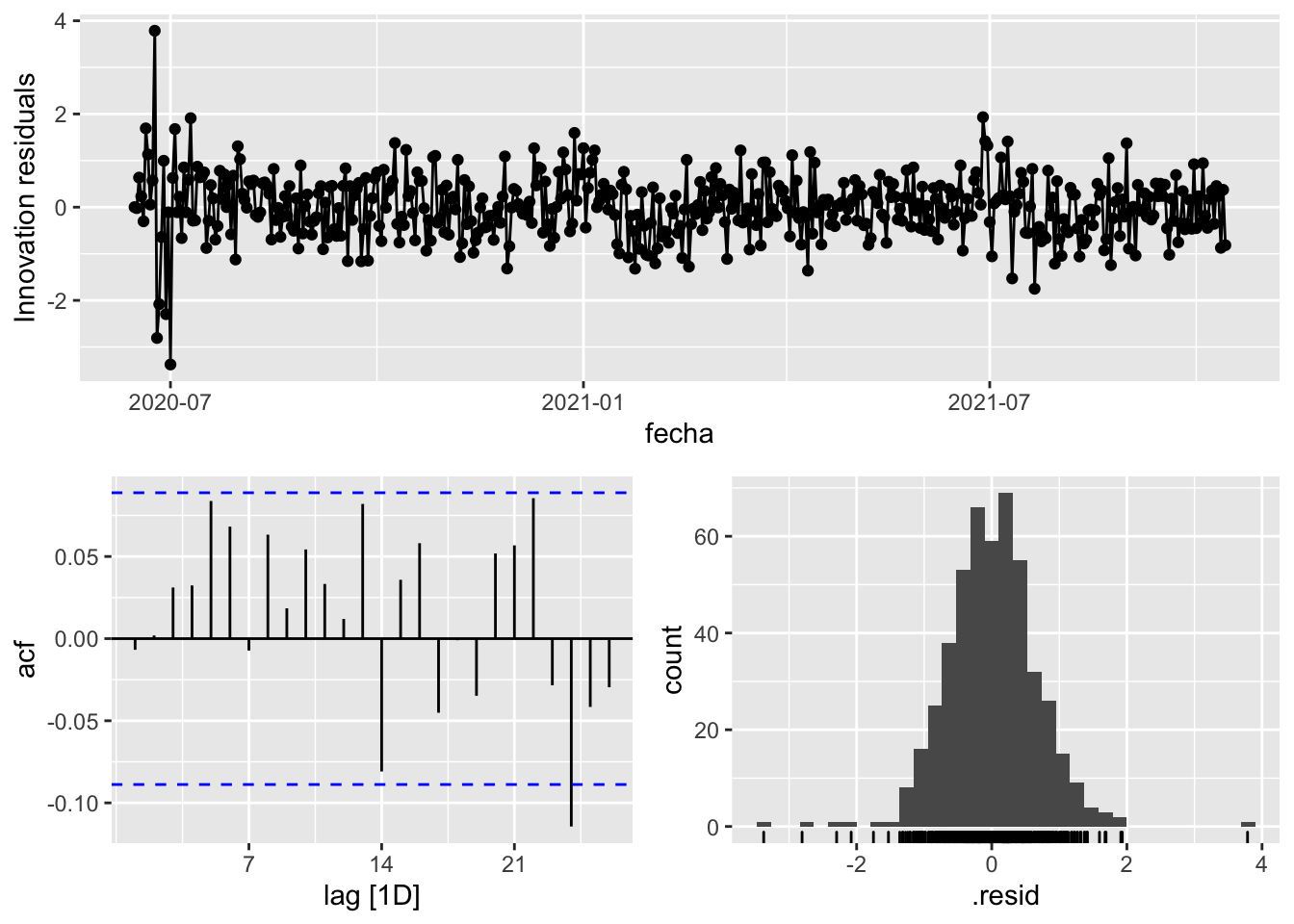
fit_model %>%
select(arima_at2) %>%
gg_tsresiduals()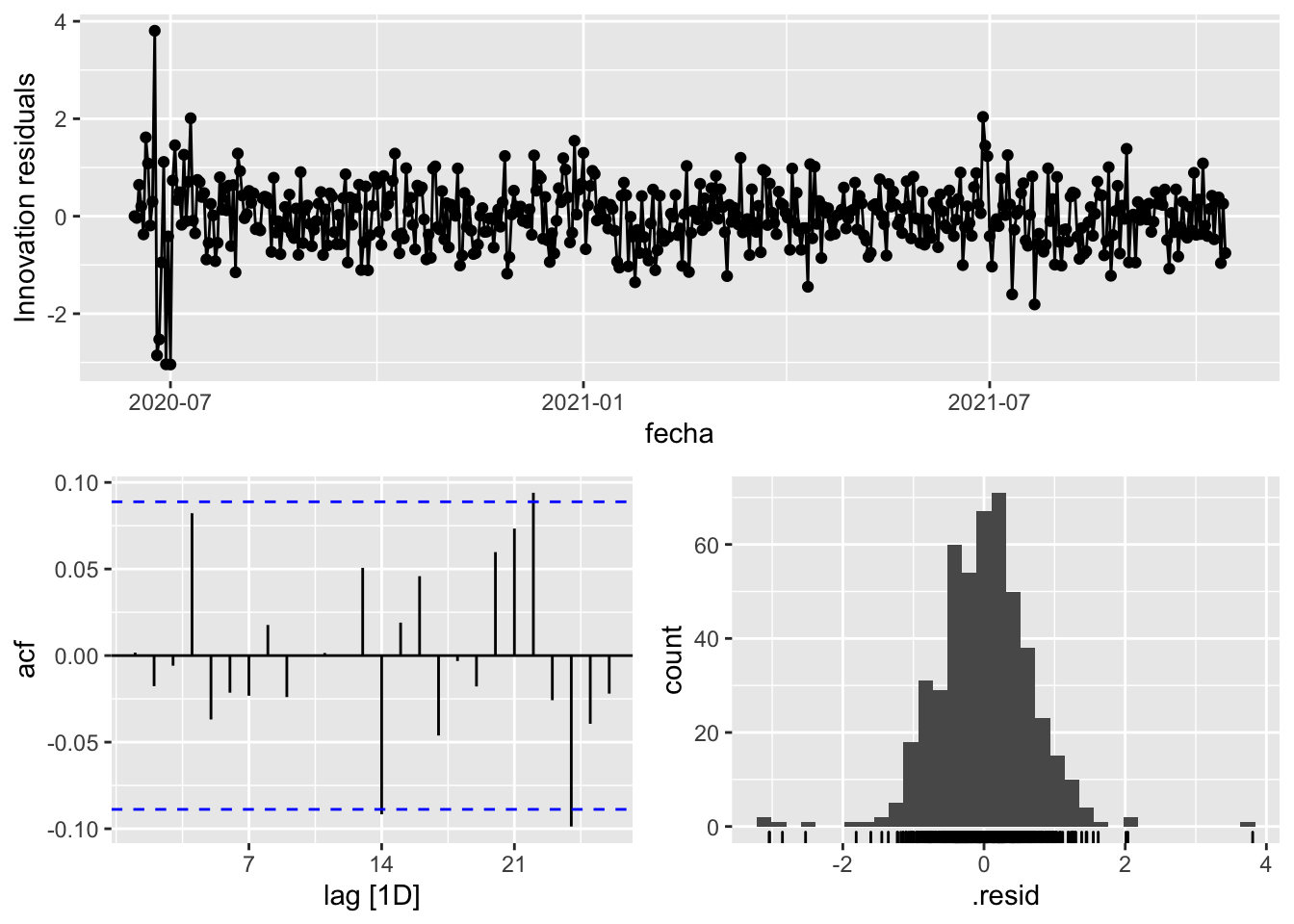
augment(fit_model) %>%
features(.innov, ljung_box, lag=7)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Málaga arima_at1 6.82 0.448
2 Málaga arima_at2 4.67 0.700
3 Málaga Snaive 1353. 0 augment(fit_model) %>%
features(.innov, ljung_box, lag=14)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Málaga arima_at1 17.7 0.219
2 Málaga arima_at2 10.6 0.715
3 Málaga Snaive 1943. 0 augment(fit_model) %>%
features(.innov, ljung_box, lag=21)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Málaga arima_at1 24.8 0.257
2 Málaga arima_at2 17.7 0.668
3 Málaga Snaive 2088. 0 augment(fit_model) %>%
features(.innov, ljung_box, lag=90)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Málaga arima_at1 121. 0.0171
2 Málaga arima_at2 87.8 0.545
3 Málaga Snaive 3170. 0 # To predict future values of infections, we need to pass the model future/prediction values of the complementary variables tmed and mobility
# We are going to select those from the test dataset
data_fc_h7 <- data_Malaga_test %>%
select(-num_casos) %>%
slice(1:7)
data_fc_h14 <- data_Malaga_test %>%
select(-num_casos) %>%
slice(1:14)
data_fc_h21 <- data_Malaga_test %>%
select(-num_casos) %>%
slice(1:21)
data_fc_h90 <- data_Malaga_test %>%
select(-num_casos) %>%
slice(1:90)# Significant spikes out of 30 is still consistent with white noise
# To be sure, use a Ljung-Box test, which has a large p-value, confirming that
# the residuals are similar to white noise.
# Note that the alternative models also passed this test.
#
# Forecast
fc_h7 <- fabletools::forecast(fit_model, new_data = data_fc_h7)
fc_h14 <- fabletools::forecast(fit_model, new_data = data_fc_h14)
fc_h21 <- fabletools::forecast(fit_model, new_data = data_fc_h21)
fc_h90 <- fabletools::forecast(fit_model, new_data = data_fc_h90)7 days
# Plots
fc_h7 %>%
autoplot(
data_Malaga_test %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(7))
) +
labs(y = "Nº cases", title = "Malaga - forecast h7")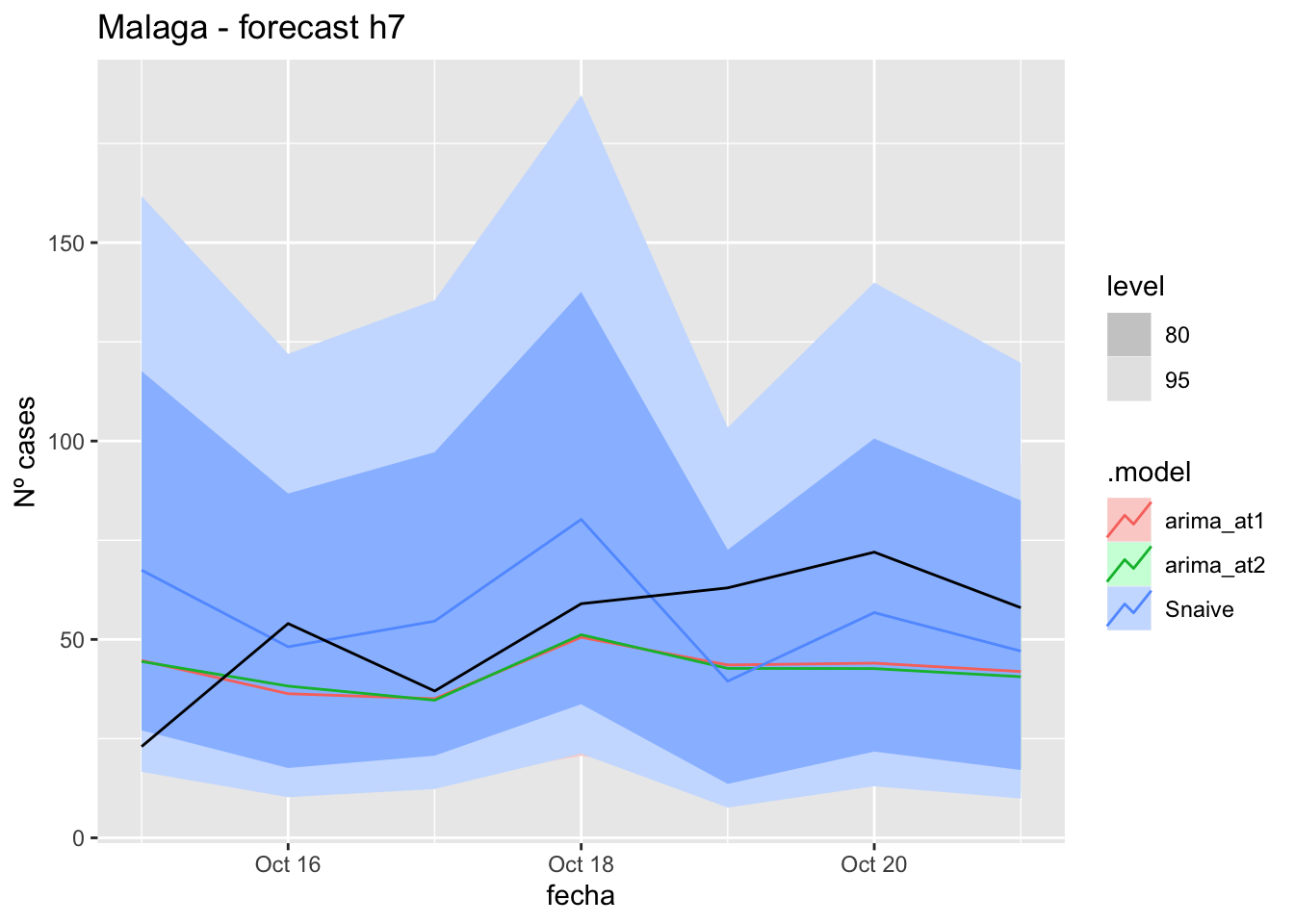
fc_h7 %>%
autoplot(
data_Malaga %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(7) &
fecha > as.Date("2021-07-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Malaga - forecast h7")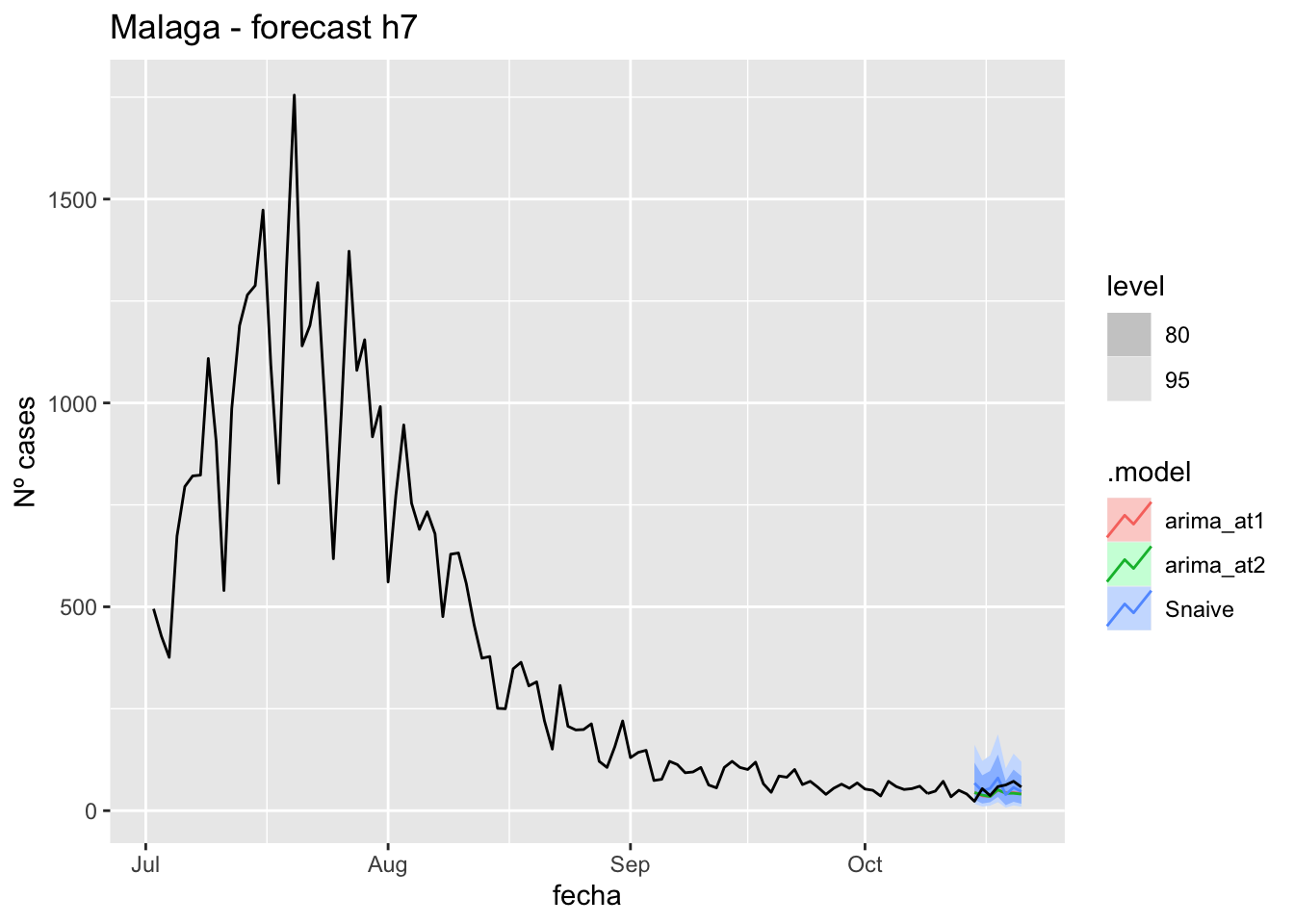
# Accuracy
fabletools::accuracy(fc_h7, data_Malaga)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_at1 Málaga Test 9.98 18.0 16.2 7.92 34.9 0.164 0.107 -0.0177
2 arima_at2 Málaga Test 10.2 18.3 16.3 8.34 35.0 0.166 0.108 0.0652
3 Snaive Málaga Test -3.97 22.9 19.8 -27.0 52.2 0.201 0.135 0.012214 days
# Plots
fc_h14 %>%
autoplot(
data_Malaga_test %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(14))
) +
labs(y = "Nº cases", title = "Malaga - forecast h14")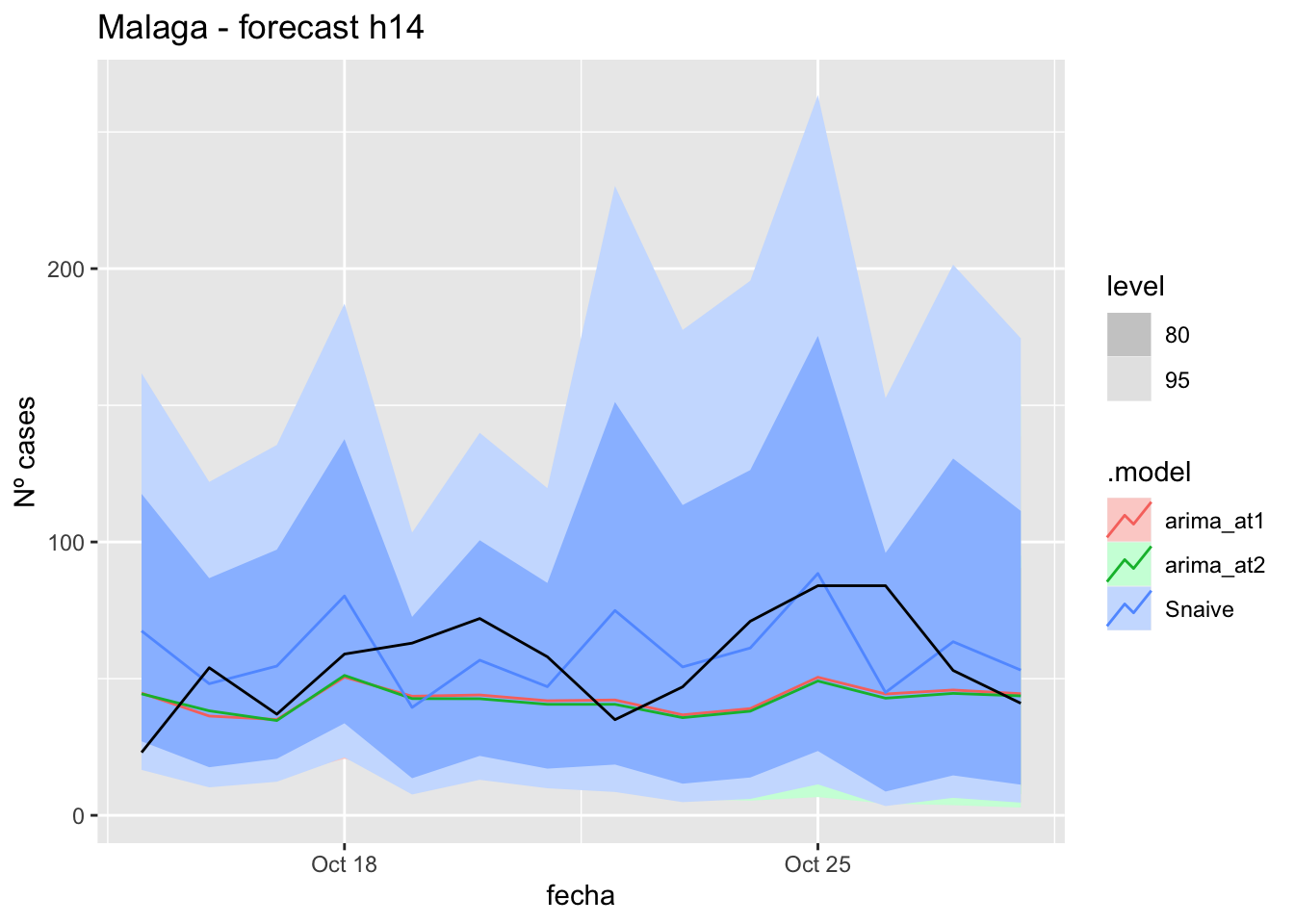
fc_h14 %>%
autoplot(
data_Malaga %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(14) &
fecha > as.Date("2021-07-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Malaga - forecast h14")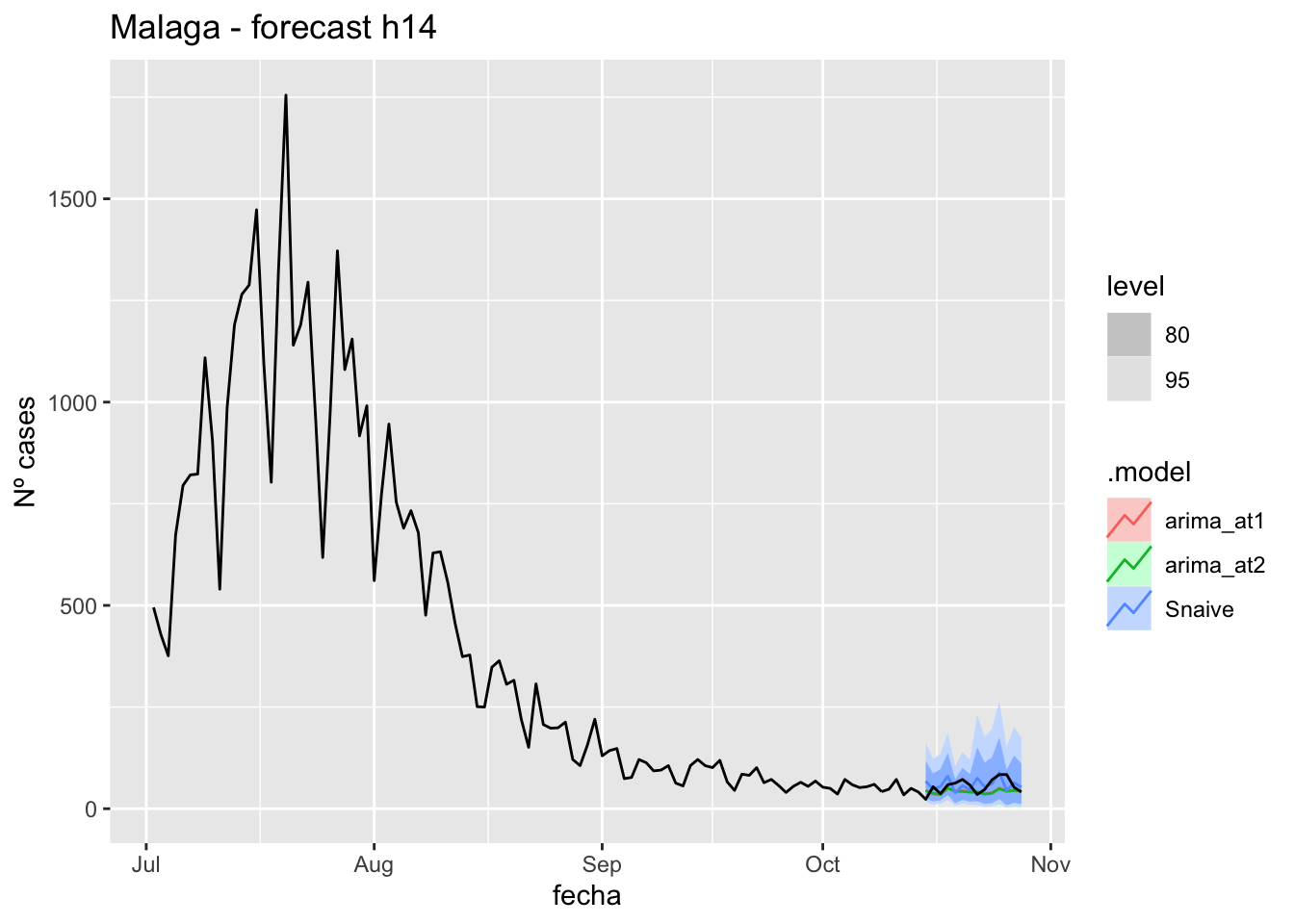
# Accuracy
fabletools::accuracy(fc_h14, data_Malaga)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_at1 Málaga Test 13.0 21.1 17.6 13.8 31.5 0.179 0.124 0.199
2 arima_at2 Málaga Test 13.7 21.7 17.9 15.2 31.7 0.182 0.128 0.241
3 Snaive Málaga Test -3.81 22.7 18.7 -22.3 43.6 0.190 0.134 -0.097521 days
# Plots
fc_h21 %>%
autoplot(
data_Malaga_test %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(21))
) +
labs(y = "Nº cases", title = "Malaga - forecast h21")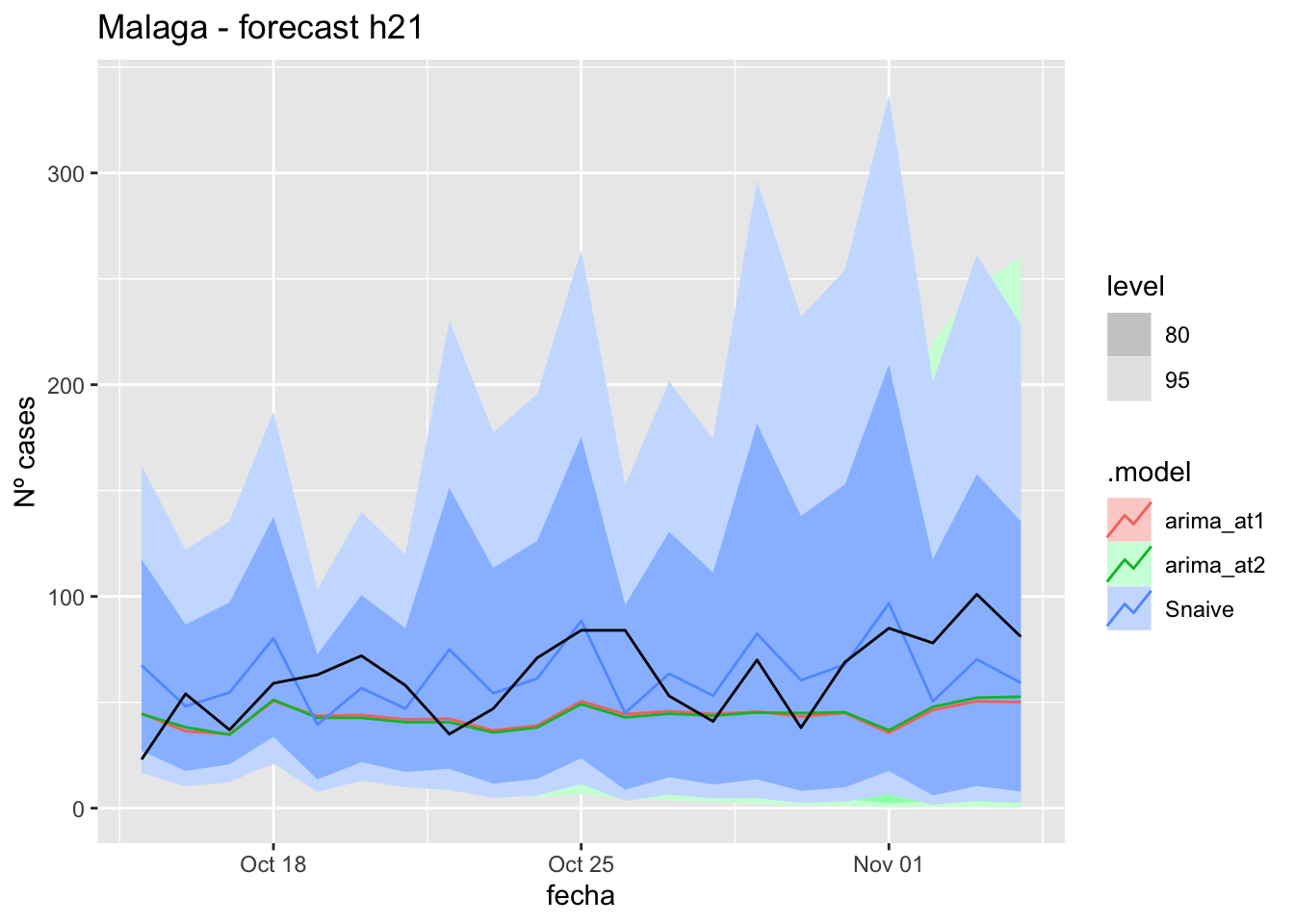
fc_h21 %>%
autoplot(
data_Malaga %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(21) &
fecha > as.Date("2021-07-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Malaga - forecast h21")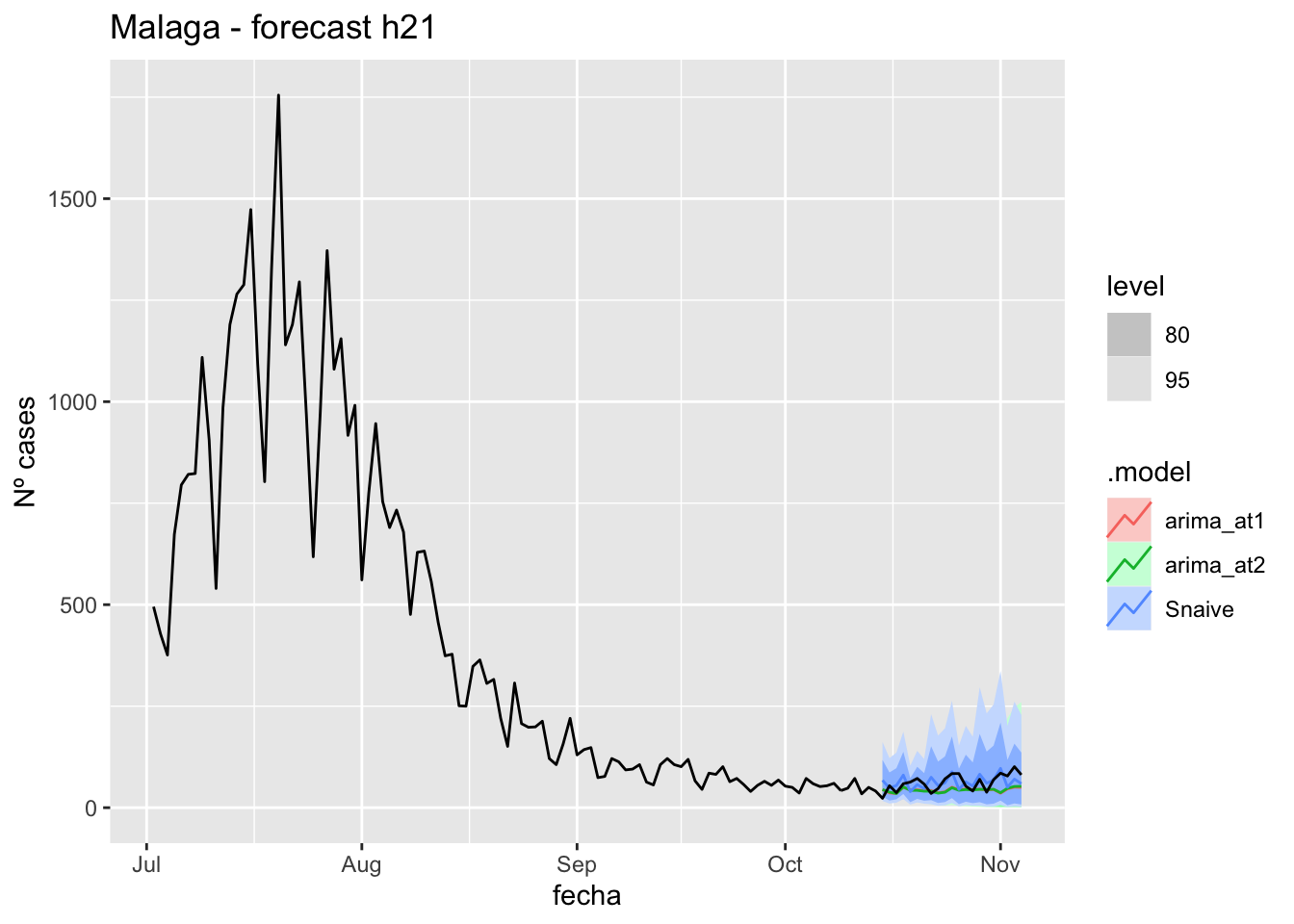
# Accuracy
fabletools::accuracy(fc_h21, data_Malaga)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_at1 Málaga Test 18.4 26.2 22.0 20.8 33.9 0.224 0.154 0.260
2 arima_at2 Málaga Test 18.5 26.0 22.0 21.1 33.8 0.223 0.154 0.246
3 Snaive Málaga Test -0.886 22.0 18.6 -14.7 37.8 0.189 0.130 0.088690 days
# Plots
fc_h90 %>%
autoplot(
data_Malaga_test %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(90))
) +
labs(y = "Nº cases", title = "Malaga - forecast h90")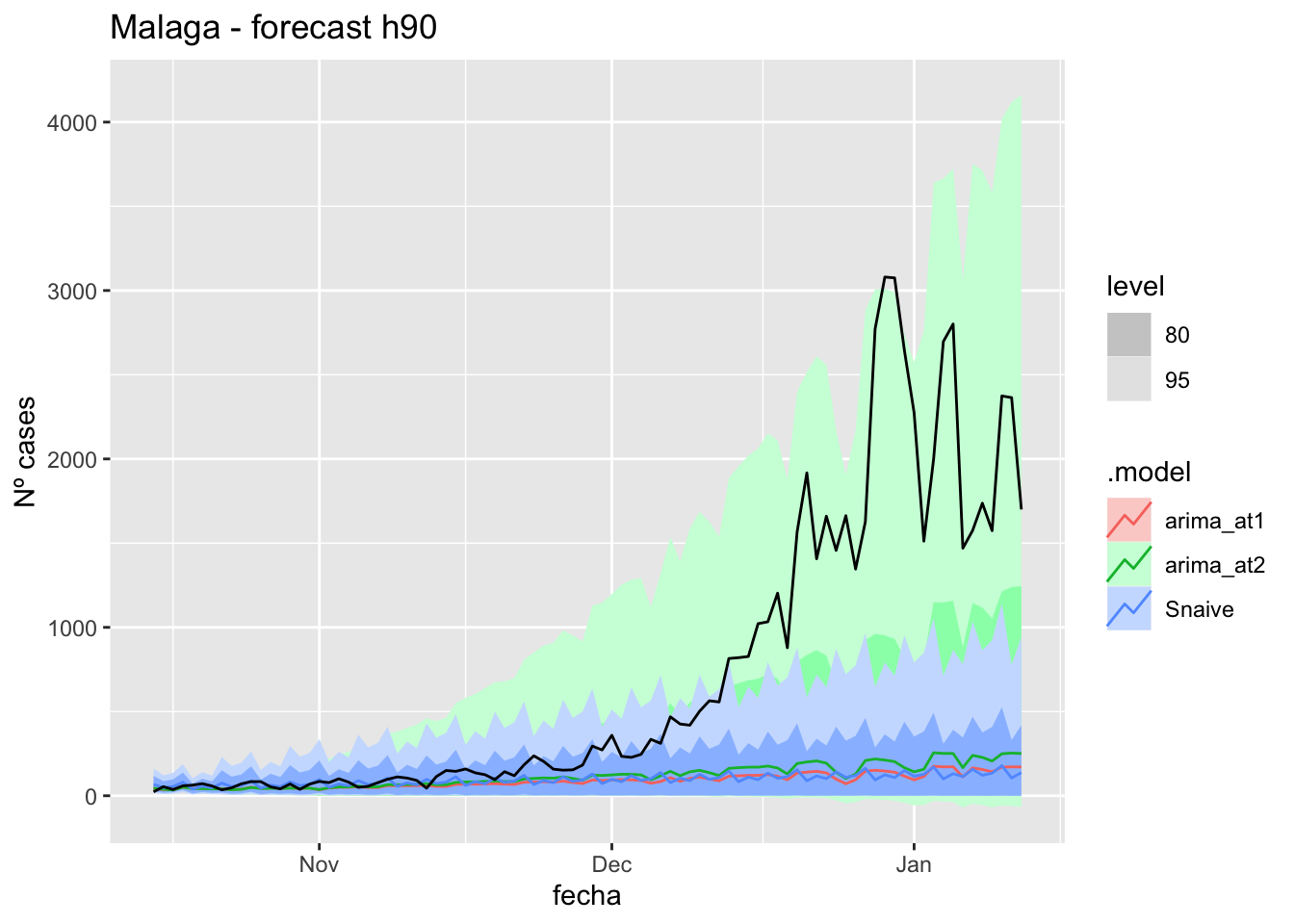
fc_h90 %>%
autoplot(
data_Malaga %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(90) &
fecha > as.Date("2021-07-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Malaga - forecast h90")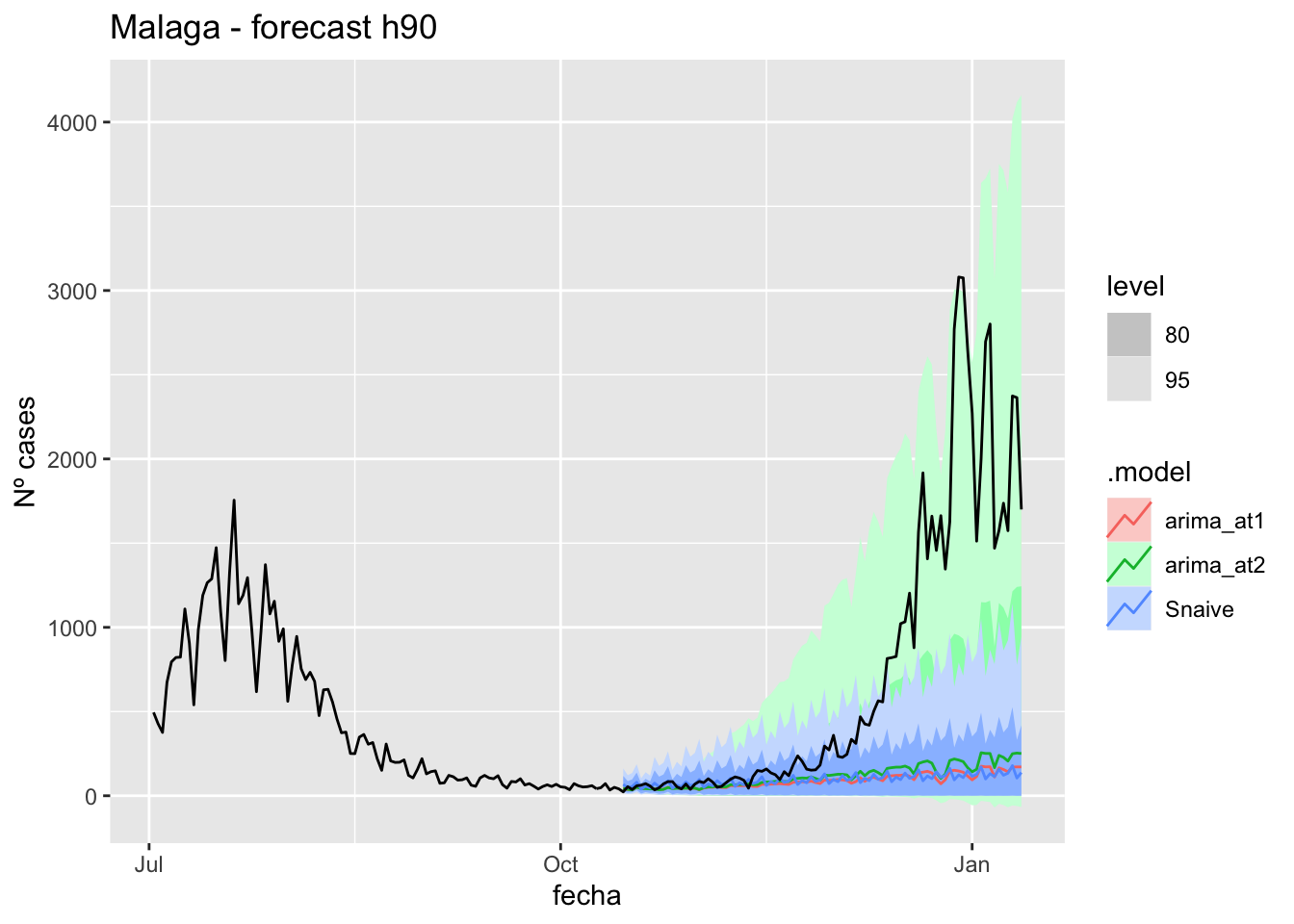
# Accuracy
fabletools::accuracy(fc_h90, data_Malaga)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_at1 Málaga Test 628. 1042. 629. 58.6 62.8 6.39 6.16 0.933
2 arima_at2 Málaga Test 599. 1007. 601. 53.2 57.8 6.10 5.95 0.932
3 Snaive Málaga Test 621. 1049. 628. 45.0 62.4 6.38 6.20 0.929Sevilla
data_Sevilla <- covid_data %>%
filter(provincia == "Sevilla") %>%
filter(fecha > as.Date("2020-06-14", format = "%Y-%m-%d")) %>%
select(provincia, fecha, num_casos, tmed, mob_grocery_pharmacy, mob_parks, mob_residential, mob_retail_recreation, mob_transit_stations, mob_workplaces) %>%
drop_na() %>%
as_tsibble(key = provincia, index = fecha)
data_Sevilla# A tsibble: 653 x 10 [1D]
# Key: provincia [1]
provincia fecha num_casos tmed mob_grocery_pharmacy mob_parks
<chr> <date> <dbl> <dbl> <dbl> <dbl>
1 Sevilla 2020-06-15 2 23.1 -14 -13
2 Sevilla 2020-06-16 1 24.0 -10 -4
3 Sevilla 2020-06-17 0 24.9 -10 -7
4 Sevilla 2020-06-18 0 25.8 -12 -13
5 Sevilla 2020-06-19 0 26.6 -13 -20
6 Sevilla 2020-06-20 0 27.5 -20 -34
7 Sevilla 2020-06-21 0 28.4 -27 -47
8 Sevilla 2020-06-22 1 29.3 -17 -19
9 Sevilla 2020-06-23 0 30.2 -12 -13
10 Sevilla 2020-06-24 1 29.4 -12 -12
# … with 643 more rows, and 4 more variables: mob_residential <dbl>,
# mob_retail_recreation <dbl>, mob_transit_stations <dbl>,
# mob_workplaces <dbl>data_Sevilla_train <- data_Sevilla %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d"))
summary(data_Sevilla_train$fecha) Min. 1st Qu. Median Mean 3rd Qu. Max.
"2020-06-15" "2020-10-14" "2021-02-13" "2021-02-13" "2021-06-14" "2021-10-14" data_Sevilla_test <- data_Sevilla %>%
filter(fecha > as.Date("2021-10-14", format = "%Y-%m-%d"))
summary(data_Sevilla_test$fecha) Min. 1st Qu. Median Mean 3rd Qu. Max.
"2021-10-15" "2021-11-25" "2022-01-05" "2022-01-05" "2022-02-15" "2022-03-29" # Lamba for num_casos
lambda_Sevilla_casos <- data_asturias %>%
features(num_casos, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Sevilla_casos[1] 0.2278229# Lamba for average temp
lambda_Sevilla_tmed <- data_asturias %>%
features(tmed, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Sevilla_tmed[1] 1.056074# Lamba for mobility
lambda_Sevilla_grocery <- data_asturias %>%
features(mob_grocery_pharmacy, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Sevilla_grocery[1] 1.377952lambda_Sevilla_parks <- data_asturias %>%
features(mob_parks, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Sevilla_parks[1] 0.7188469lambda_Sevilla_resd <- data_asturias %>%
features(mob_residential, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Sevilla_resd[1] 1.000064lambda_Sevilla_retail <- data_asturias %>%
features(mob_retail_recreation, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Sevilla_retail[1] 1.058286lambda_Sevilla_transit <- data_asturias %>%
features(mob_transit_stations, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Sevilla_transit[1] 1.116829lambda_Sevilla_work <- data_asturias %>%
features(mob_workplaces, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Sevilla_work[1] 1.999927fit_model <- data_Sevilla_train %>%
model(
arima_at1 = ARIMA(box_cox(num_casos, lambda_Sevilla_casos) ~
box_cox(tmed, lambda_Sevilla_tmed) +
box_cox(mob_grocery_pharmacy, lambda_Sevilla_grocery) +
box_cox(mob_parks, lambda_Sevilla_parks) +
box_cox(mob_residential, lambda_Sevilla_resd) +
box_cox(mob_retail_recreation, lambda_Sevilla_retail) +
box_cox(mob_transit_stations, lambda_Sevilla_transit) +
box_cox(mob_workplaces, lambda_Sevilla_work)
),
arima_at2 = ARIMA(box_cox(num_casos, lambda_Sevilla_casos) ~
box_cox(tmed, lambda_Sevilla_tmed) +
box_cox(mob_grocery_pharmacy, lambda_Sevilla_grocery) +
box_cox(mob_parks, lambda_Sevilla_parks) +
box_cox(mob_residential, lambda_Sevilla_resd) +
box_cox(mob_retail_recreation, lambda_Sevilla_retail) +
box_cox(mob_transit_stations, lambda_Sevilla_transit) +
box_cox(mob_workplaces, lambda_Sevilla_work),
stepwise = FALSE, approx = FALSE),
Snaive = SNAIVE(box_cox(num_casos, lambda_Sevilla_casos))
)
# Show and report model
fit_model# A mable: 1 x 4
# Key: provincia [1]
provincia arima_at1
<chr> <model>
1 Sevilla <LM w/ ARIMA(3,1,1)(2,0,0)[7] errors>
# … with 2 more variables: arima_at2 <model>, Snaive <model>glance(fit_model)# A tibble: 3 × 9
provincia .model sigma2 log_lik AIC AICc BIC ar_roots ma_roots
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <list> <list>
1 Sevilla arima_at1 0.855 -647. 1321. 1322. 1380. <cpl [17]> <cpl [1]>
2 Sevilla arima_at2 0.837 -643. 1311. 1312. 1366. <cpl [10]> <cpl [7]>
3 Sevilla Snaive 2.47 NA NA NA NA <NULL> <NULL> fit_model %>%
pivot_longer(-provincia,
names_to = ".model",
values_to = "orders") %>%
left_join(glance(fit_model), by = c("provincia", ".model")) %>%
arrange(AICc) %>%
select(.model:BIC)# A mable: 3 x 7
# Key: .model [3]
.model orders sigma2 log_lik AIC AICc BIC
<chr> <model> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_… <LM w/ ARIMA(3,1,0)(1,0,1)[7] errors> 0.837 -643. 1311. 1312. 1366.
2 arima_… <LM w/ ARIMA(3,1,1)(2,0,0)[7] errors> 0.855 -647. 1321. 1322. 1380.
3 Snaive <SNAIVE> 2.47 NA NA NA NA # We use a Ljung-Box test >> large p-value, confirms residuals are similar / considered to white noise
fit_model %>%
select(Snaive) %>%
gg_tsresiduals()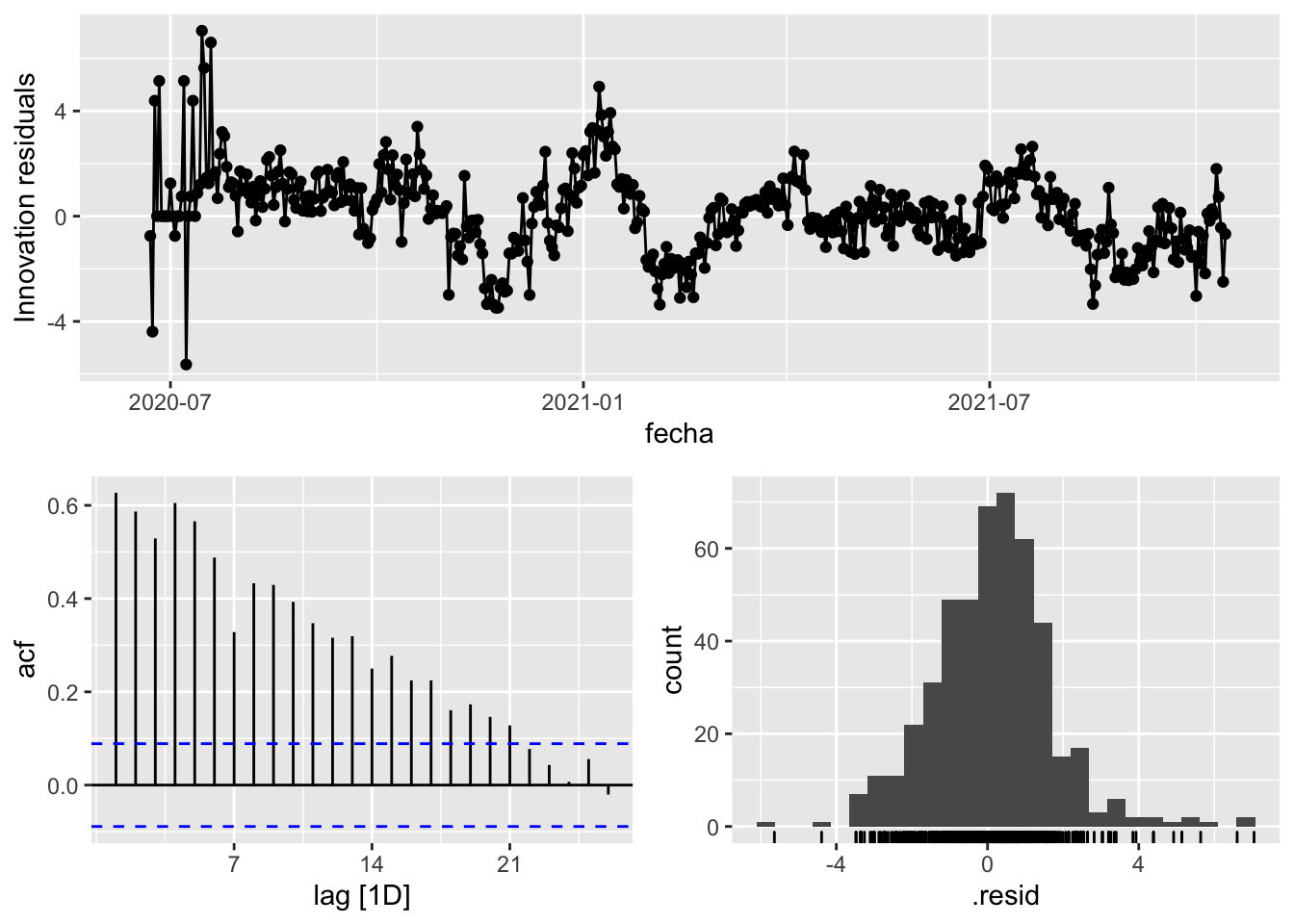
fit_model %>%
select(arima_at1) %>%
gg_tsresiduals()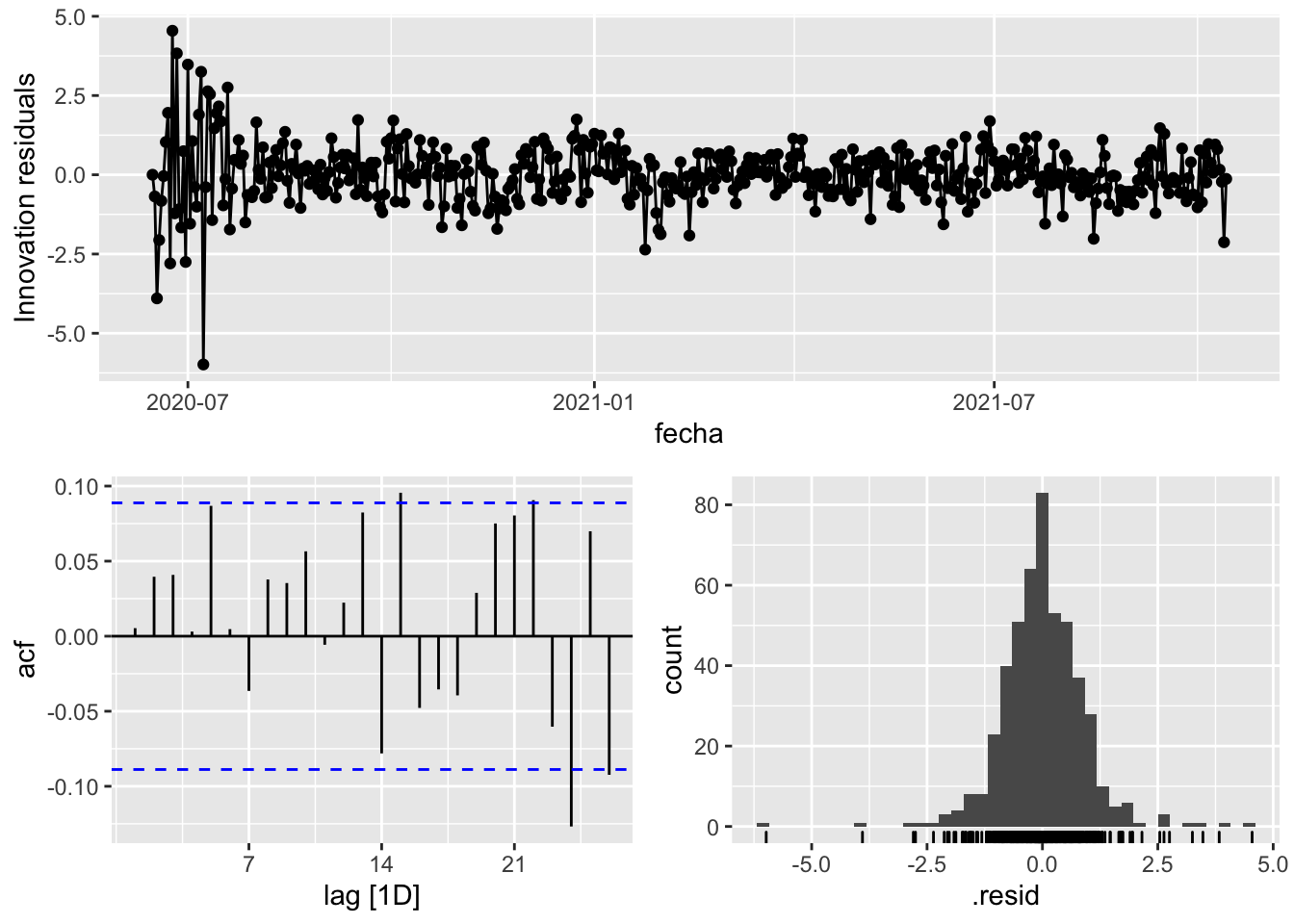
fit_model %>%
select(arima_at2) %>%
gg_tsresiduals()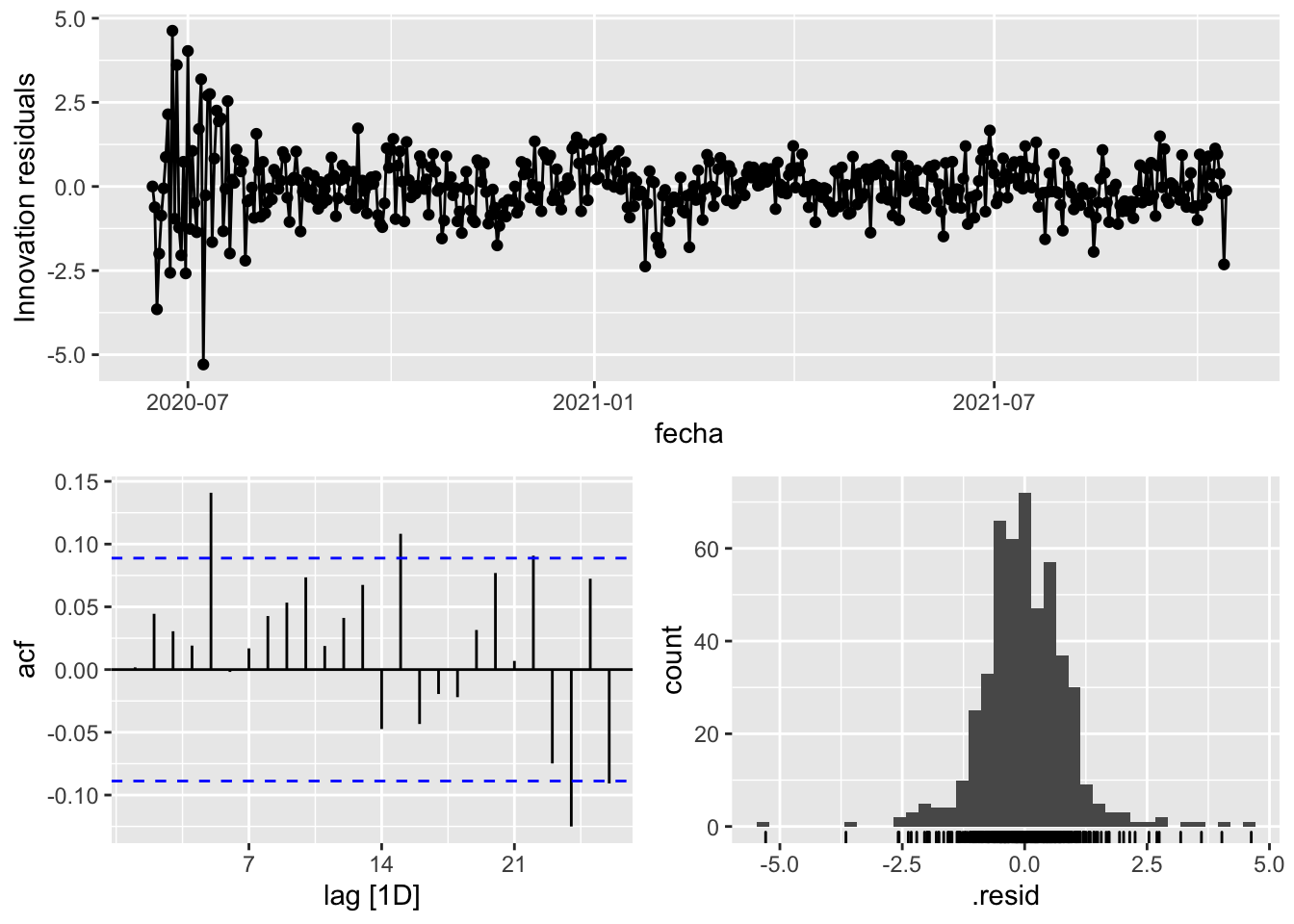
augment(fit_model) %>%
features(.innov, ljung_box, lag=7)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Sevilla arima_at1 6.00 0.539
2 Sevilla arima_at2 11.6 0.116
3 Sevilla Snaive 995. 0 augment(fit_model) %>%
features(.innov, ljung_box, lag=14)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Sevilla arima_at1 15.7 0.333
2 Sevilla arima_at2 21.0 0.101
3 Sevilla Snaive 1444. 0 augment(fit_model) %>%
features(.innov, ljung_box, lag=21)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Sevilla arima_at1 29.5 0.103
2 Sevilla arima_at2 31.9 0.0603
3 Sevilla Snaive 1580. 0 augment(fit_model) %>%
features(.innov, ljung_box, lag=90)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Sevilla arima_at1 124. 0.0104
2 Sevilla arima_at2 145. 0.000218
3 Sevilla Snaive 2311. 0 # To predict future values of infections, we need to pass the model future/prediction values of the complementary variables tmed and mobility
# We are going to select those from the test dataset
data_fc_h7 <- data_Sevilla_test %>%
select(-num_casos) %>%
slice(1:7)
data_fc_h14 <- data_Sevilla_test %>%
select(-num_casos) %>%
slice(1:14)
data_fc_h21 <- data_Sevilla_test %>%
select(-num_casos) %>%
slice(1:21)
data_fc_h90 <- data_Sevilla_test %>%
select(-num_casos) %>%
slice(1:90)# Significant spikes out of 30 is still consistent with white noise
# To be sure, use a Ljung-Box test, which has a large p-value, confirming that
# the residuals are similar to white noise.
# Note that the alternative models also passed this test.
#
# Forecast
fc_h7 <- fabletools::forecast(fit_model, new_data = data_fc_h7)
fc_h14 <- fabletools::forecast(fit_model, new_data = data_fc_h14)
fc_h21 <- fabletools::forecast(fit_model, new_data = data_fc_h21)
fc_h90 <- fabletools::forecast(fit_model, new_data = data_fc_h90)7 days
# Plots
fc_h7 %>%
autoplot(
data_Sevilla_test %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(7))
) +
labs(y = "Nº cases", title = "Sevilla - forecast h7")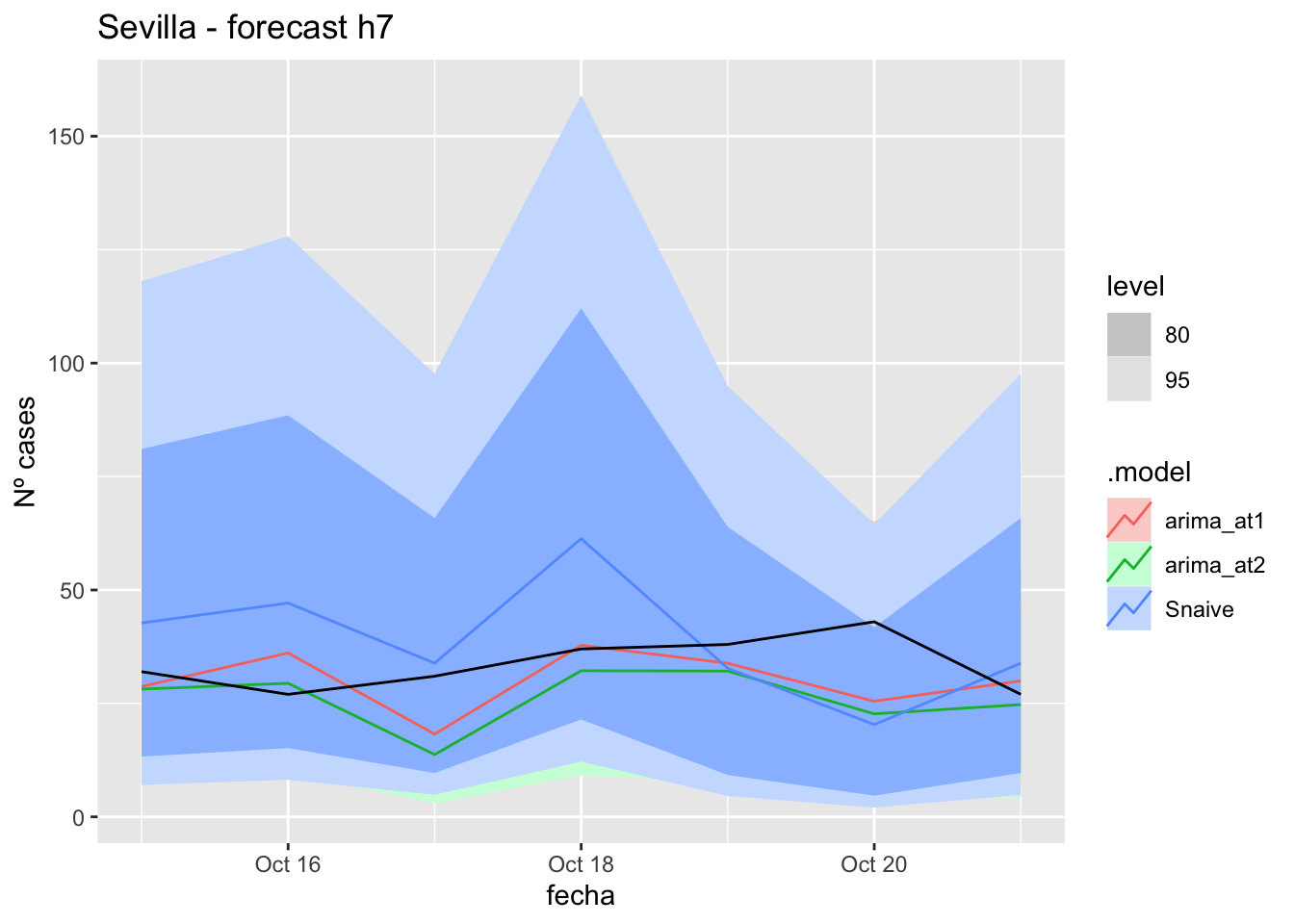
fc_h7 %>%
autoplot(
data_Sevilla %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(7) &
fecha > as.Date("2021-07-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Sevilla - forecast h7")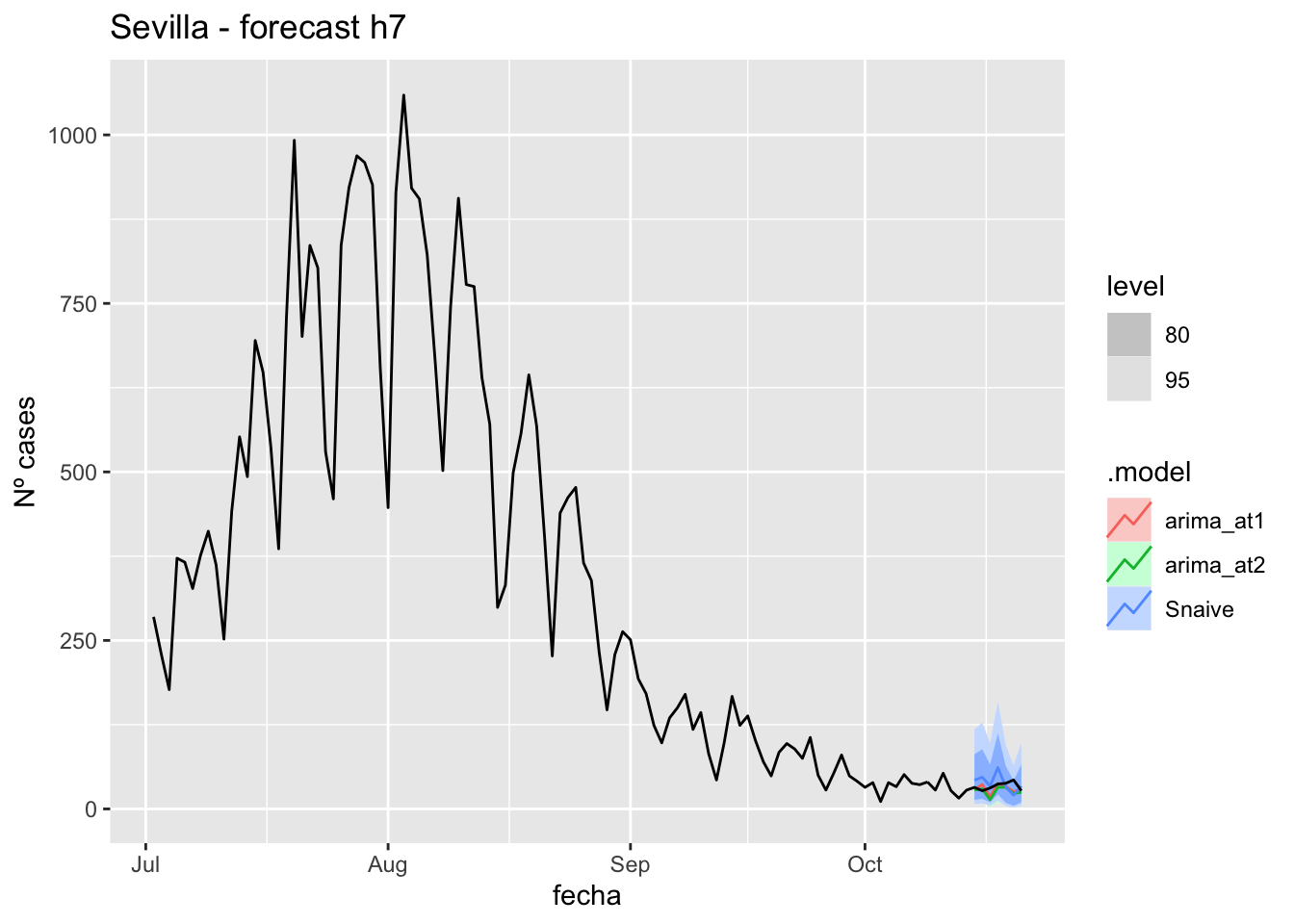
# Accuracy
fabletools::accuracy(fc_h7, data_Sevilla)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_at1 Sevilla Test 3.54 9.18 7.23 8.02 21.4 0.0714 0.0582 -0.475
2 arima_at2 Sevilla Test 7.41 10.6 8.11 20.4 23.0 0.0801 0.0675 -0.416
3 Snaive Sevilla Test -5.28 15.6 13.3 -20.3 39.3 0.131 0.0990 0.032314 days
# Plots
fc_h14 %>%
autoplot(
data_Sevilla_test %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(14))
) +
labs(y = "Nº cases", title = "Sevilla - forecast h14")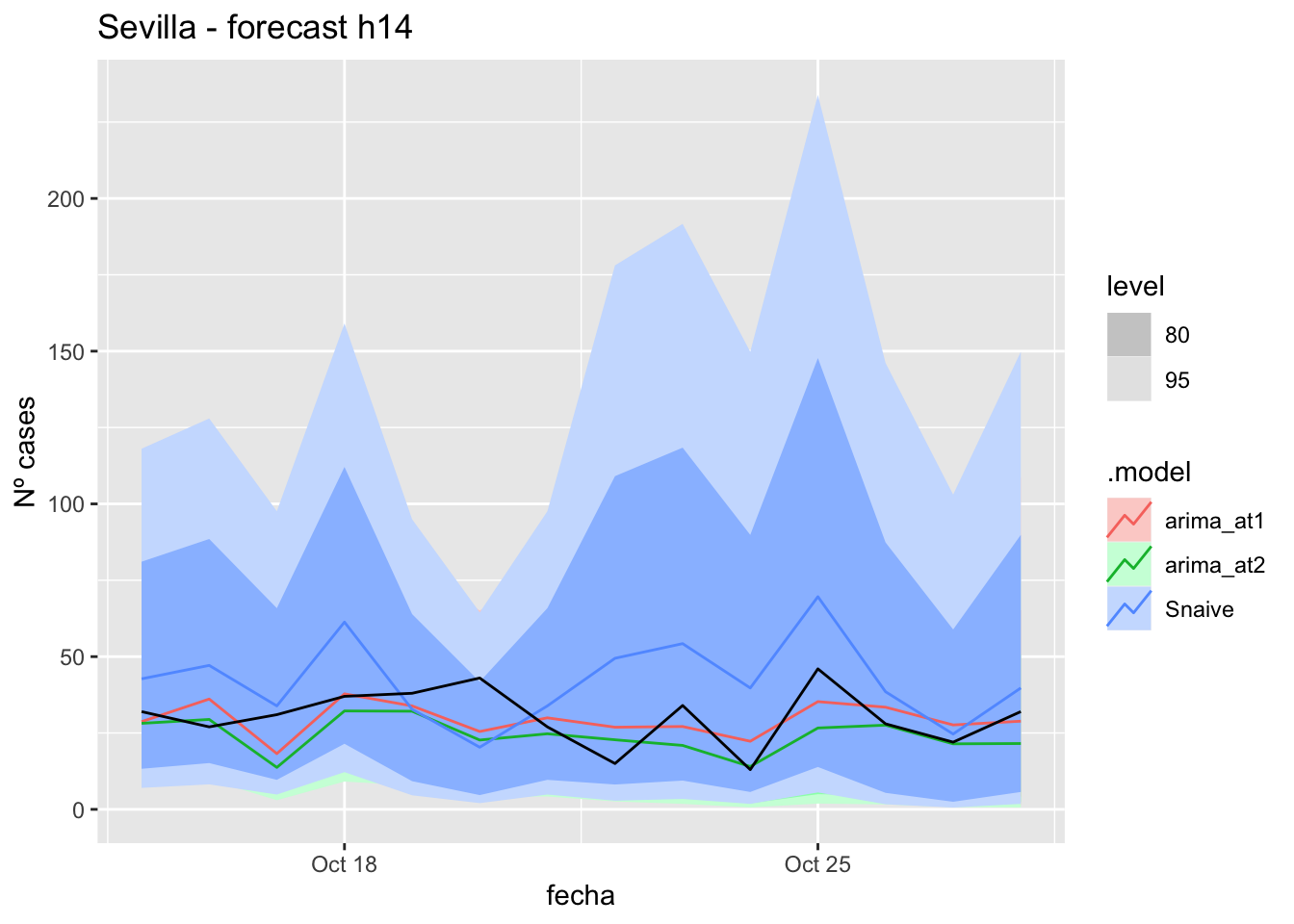
fc_h14 %>%
autoplot(
data_Sevilla %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(14) &
fecha > as.Date("2021-07-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Sevilla - forecast h14")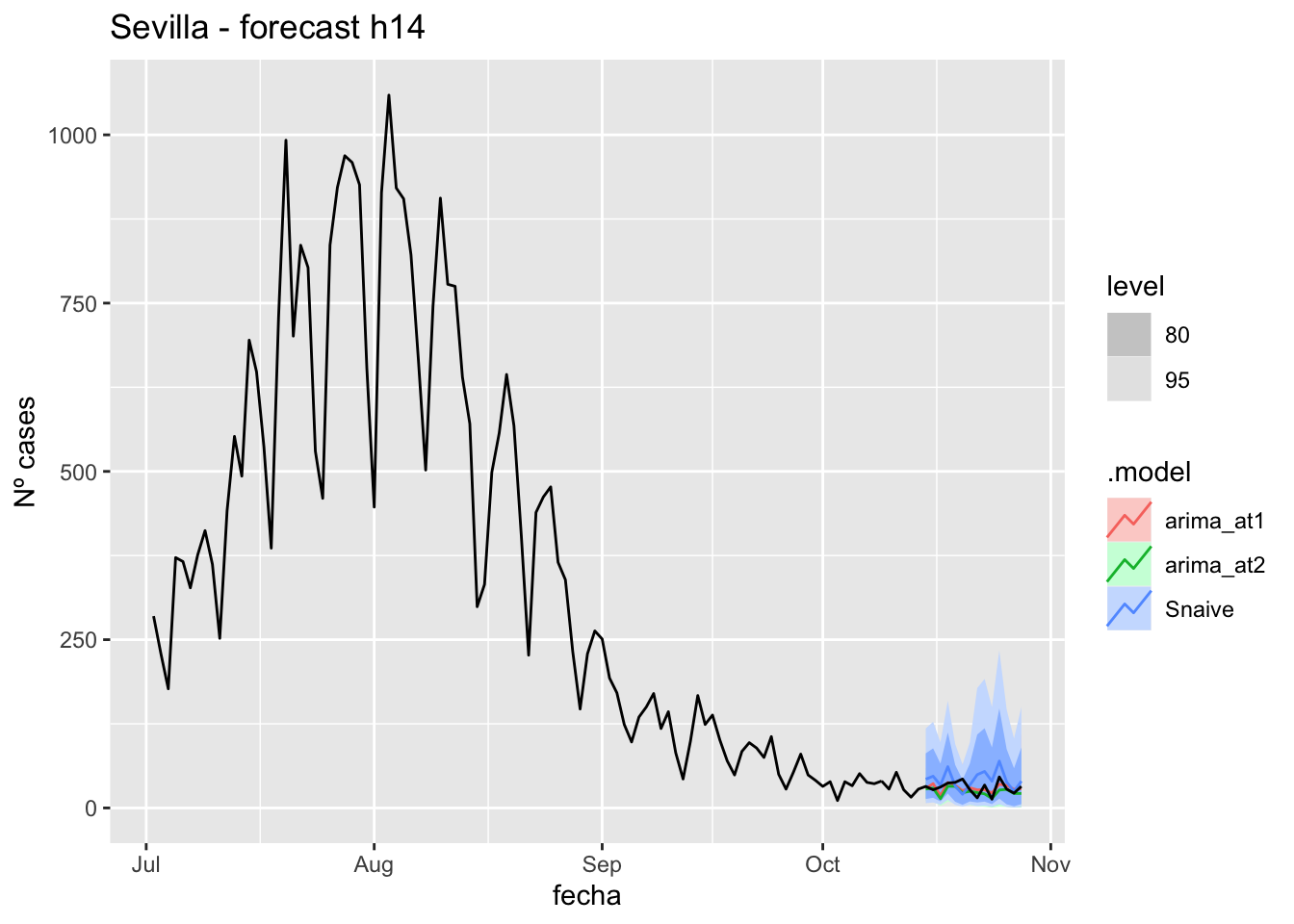
# Accuracy
fabletools::accuracy(fc_h14, data_Sevilla)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_a… Sevilla Test 0.949 8.67 7.40 -6.17 28.5 0.0731 0.0549 -0.387
2 arima_a… Sevilla Test 6.21 10.4 7.82 14.3 24.1 0.0772 0.0659 -0.416
3 Snaive Sevilla Test -11.6 18.4 15.6 -54.4 63.9 0.154 0.117 0.26821 days
# Plots
fc_h21 %>%
autoplot(
data_Sevilla_test %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(21))
) +
labs(y = "Nº cases", title = "Sevilla - forecast h21")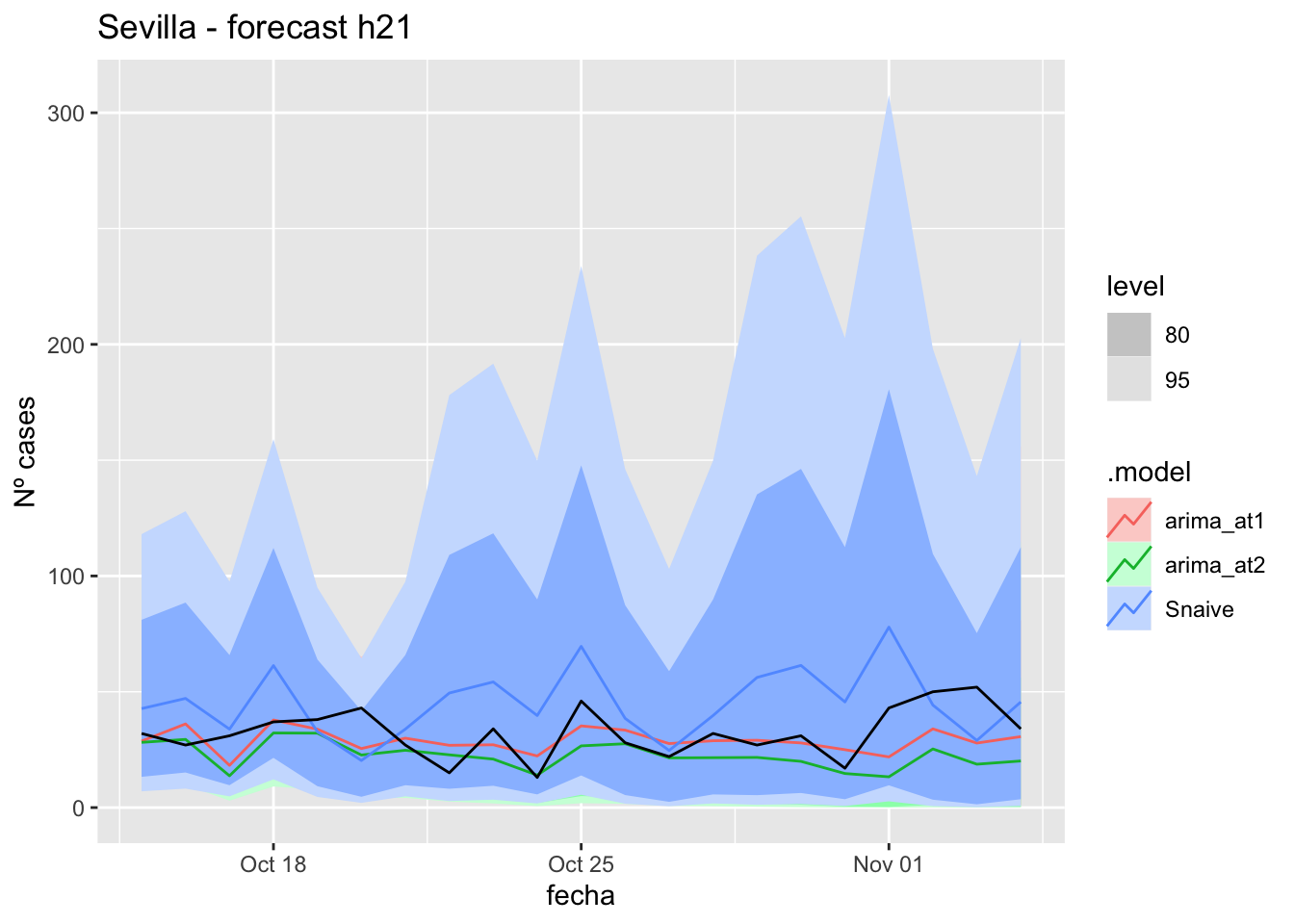
fc_h21 %>%
autoplot(
data_Sevilla %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(21) &
fecha > as.Date("2021-07-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Sevilla - forecast h21")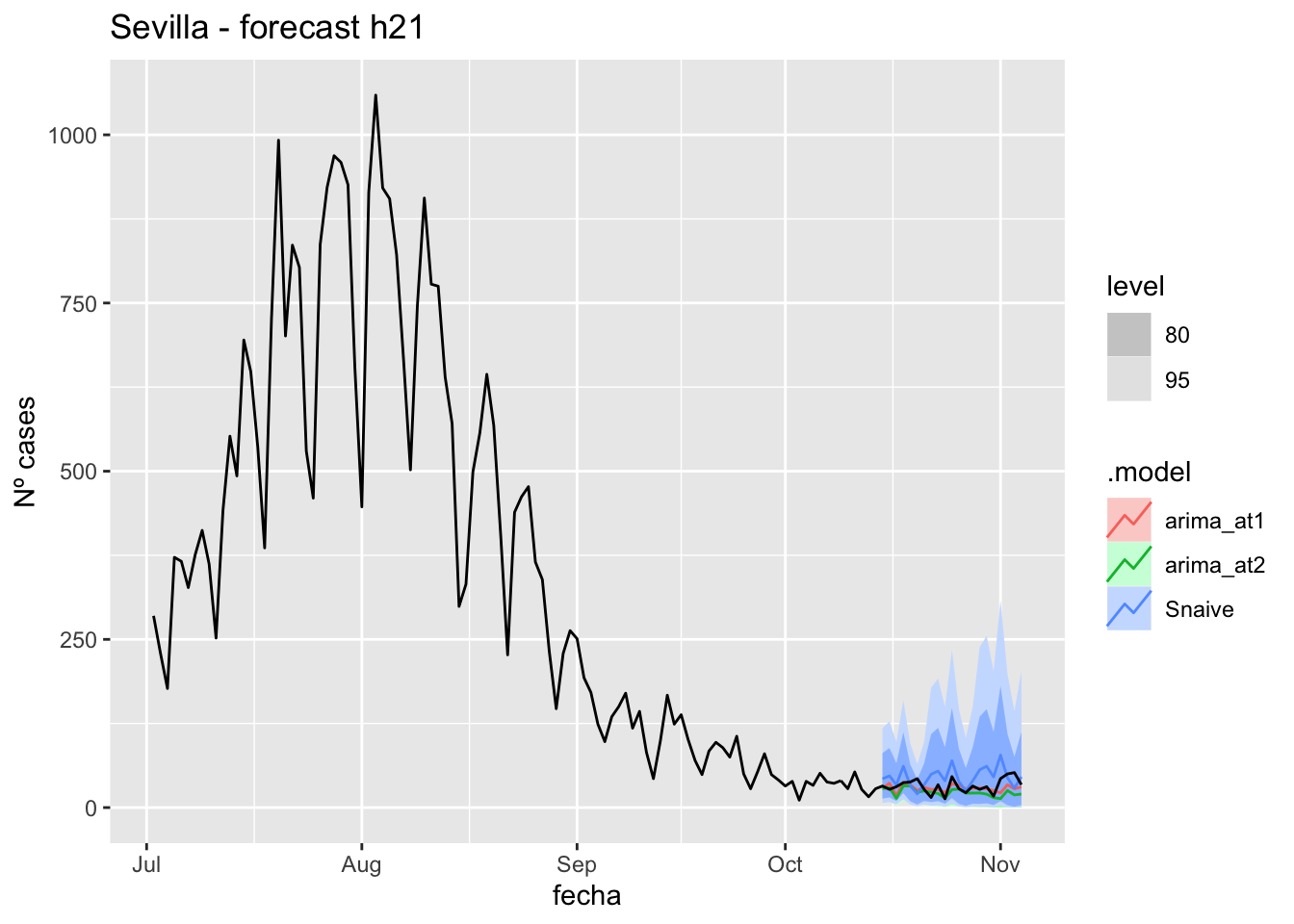
# Accuracy
fabletools::accuracy(fc_h21, data_Sevilla)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_… Sevilla Test 3.38 10.7 8.64 0.302 28.6 0.0853 0.0681 -0.0110
2 arima_… Sevilla Test 9.87 14.6 10.9 23.4 30.0 0.108 0.0924 0.143
3 Snaive Sevilla Test -12.8 21.0 18.2 -57.0 68.6 0.180 0.133 0.328 90 days
# Plots
fc_h90 %>%
autoplot(
data_Sevilla_test %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(90))
) +
labs(y = "Nº cases", title = "Sevilla - forecast h90")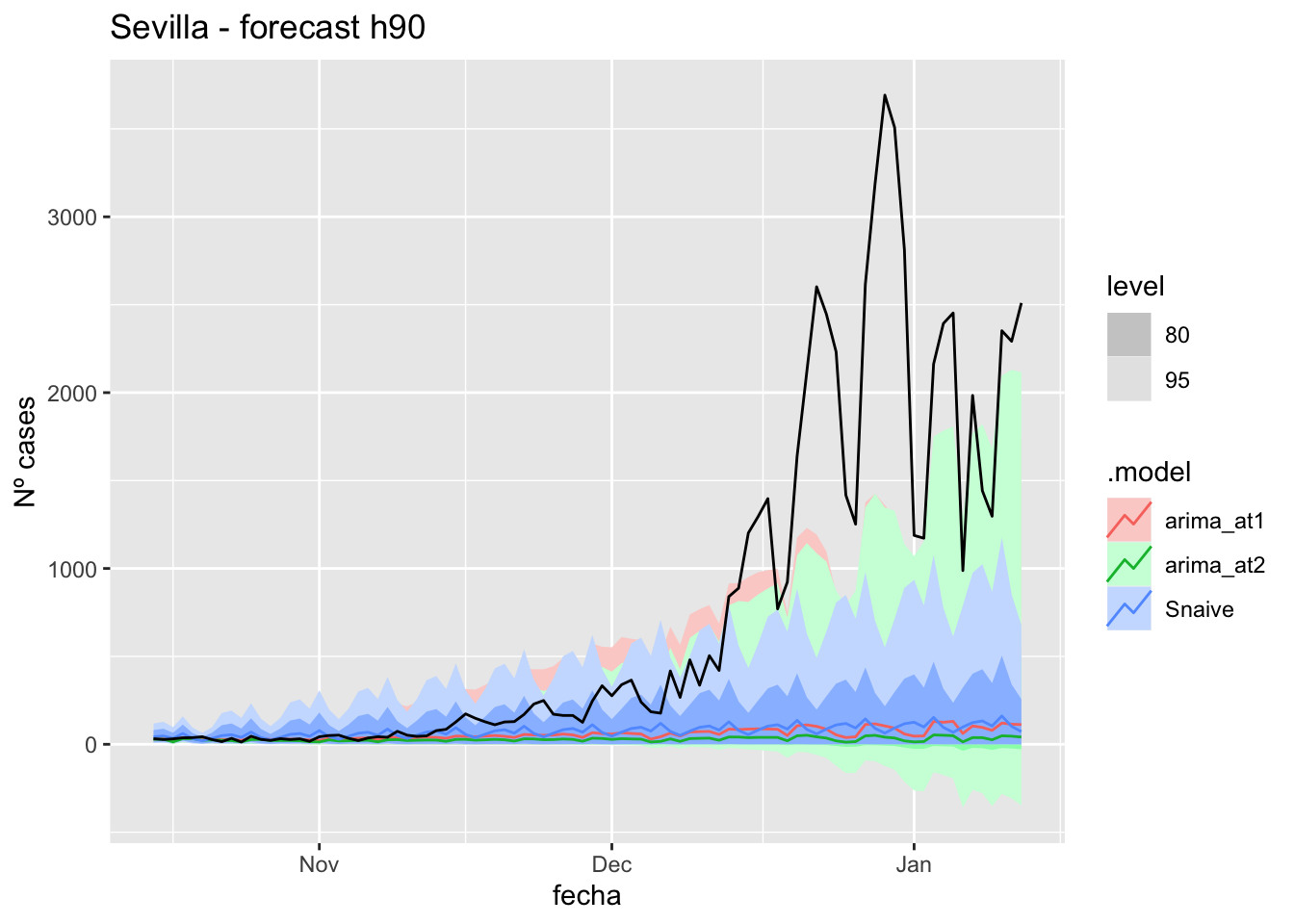
fc_h90 %>%
autoplot(
data_Sevilla %>%
filter(fecha <= as.Date("2021-10-14", format = "%Y-%m-%d")+days(90) &
fecha > as.Date("2021-07-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Sevilla - forecast h90")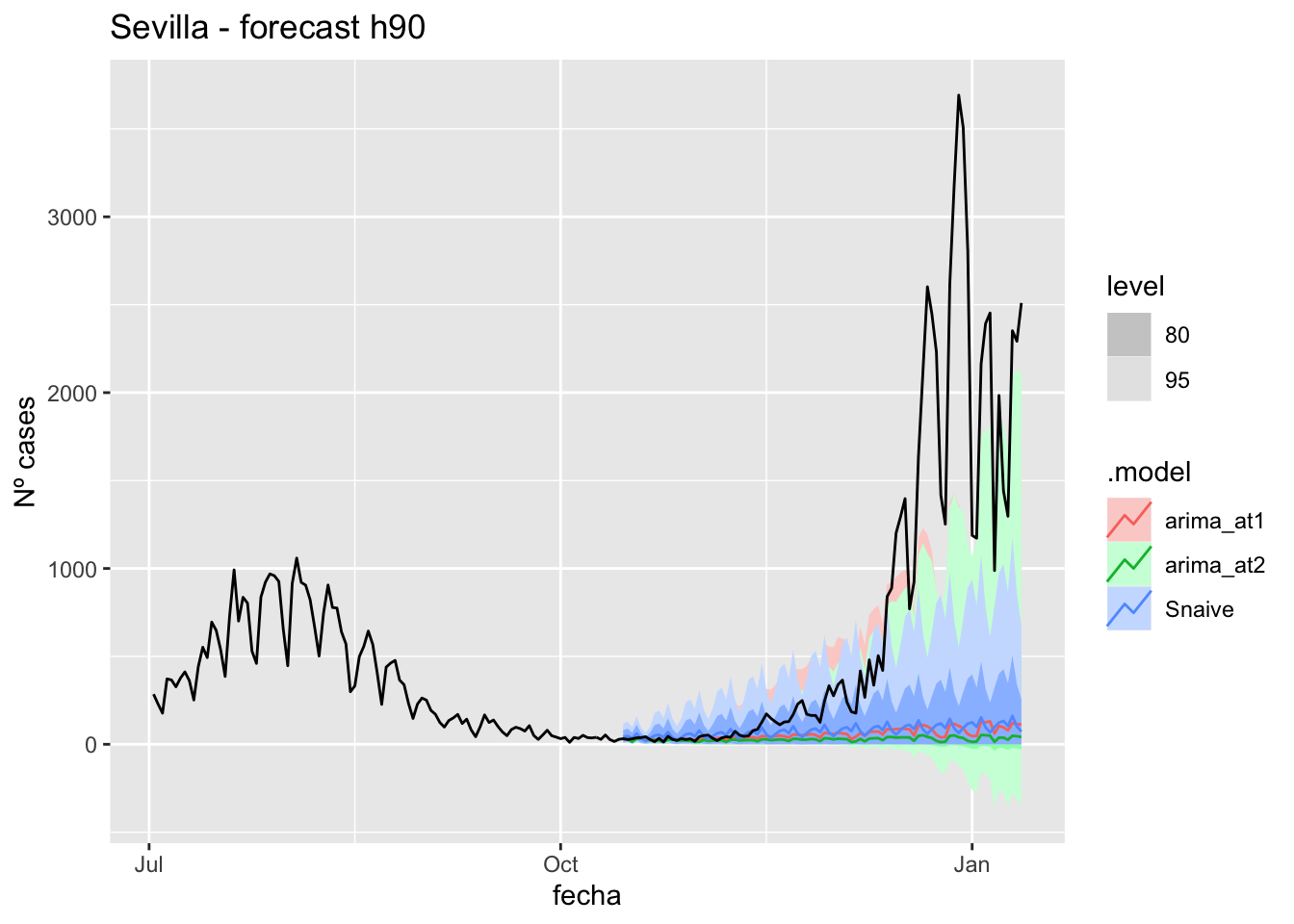
# Accuracy
fabletools::accuracy(fc_h90, data_Sevilla)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_at1 Sevilla Test 687. 1168. 689. 58.0 66.1 6.80 7.40 0.898
2 arima_at2 Sevilla Test 717. 1200. 717. 71.8 73.5 7.09 7.61 0.898
3 Snaive Sevilla Test 669. 1164. 680. 34.0 74.1 6.72 7.38 0.893Middel of sixth epidemiological period
Asturias
data_asturias <- covid_data %>%
filter(provincia == "Asturias") %>%
filter(fecha > as.Date("2020-06-14", format = "%Y-%m-%d")) %>%
select(provincia, fecha, num_casos, tmed, mob_grocery_pharmacy, mob_parks, mob_residential, mob_retail_recreation, mob_transit_stations, mob_workplaces) %>%
drop_na() %>%
as_tsibble(key = provincia, index = fecha)
data_asturias# A tsibble: 653 x 10 [1D]
# Key: provincia [1]
provincia fecha num_casos tmed mob_grocery_pharmacy mob_parks
<chr> <date> <dbl> <dbl> <dbl> <dbl>
1 Asturias 2020-06-15 0 16 -6 39
2 Asturias 2020-06-16 1 15.6 -6 7
3 Asturias 2020-06-17 0 15.6 -7 20
4 Asturias 2020-06-18 0 15.1 -4 68
5 Asturias 2020-06-19 0 14.8 -4 52
6 Asturias 2020-06-20 0 16.8 -10 71
7 Asturias 2020-06-21 0 18.2 -5.5 46
8 Asturias 2020-06-22 0 18.2 -1 99
9 Asturias 2020-06-23 0 20 -2 129
10 Asturias 2020-06-24 0 18.2 -9 63
# … with 643 more rows, and 4 more variables: mob_residential <dbl>,
# mob_retail_recreation <dbl>, mob_transit_stations <dbl>,
# mob_workplaces <dbl>data_asturias_train <- data_asturias %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d"))
summary(data_asturias_train$fecha) Min. 1st Qu. Median Mean 3rd Qu. Max.
"2020-06-15" "2020-11-10" "2021-04-08" "2021-04-08" "2021-09-04" "2022-01-31" data_asturias_test <- data_asturias %>%
filter(fecha > as.Date("2022-01-31", format = "%Y-%m-%d"))
summary(data_asturias_test$fecha) Min. 1st Qu. Median Mean 3rd Qu. Max.
"2022-02-01" "2022-02-15" "2022-03-01" "2022-03-01" "2022-03-15" "2022-03-29" # Lamba for num_casos
lambda_asturias_casos <- data_asturias %>%
features(num_casos, features = guerrero) %>%
pull(lambda_guerrero)
lambda_asturias_casos[1] 0.2278229# Lamba for average temp
lambda_asturias_tmed <- data_asturias %>%
features(tmed, features = guerrero) %>%
pull(lambda_guerrero)
lambda_asturias_tmed[1] 1.056074# Lamba for mobility
lambda_asturias_grocery <- data_asturias %>%
features(mob_grocery_pharmacy, features = guerrero) %>%
pull(lambda_guerrero)
lambda_asturias_grocery[1] 1.377952lambda_asturias_parks <- data_asturias %>%
features(mob_parks, features = guerrero) %>%
pull(lambda_guerrero)
lambda_asturias_parks[1] 0.7188469lambda_asturias_resd <- data_asturias %>%
features(mob_residential, features = guerrero) %>%
pull(lambda_guerrero)
lambda_asturias_resd[1] 1.000064lambda_asturias_retail <- data_asturias %>%
features(mob_retail_recreation, features = guerrero) %>%
pull(lambda_guerrero)
lambda_asturias_retail[1] 1.058286lambda_asturias_transit <- data_asturias %>%
features(mob_transit_stations, features = guerrero) %>%
pull(lambda_guerrero)
lambda_asturias_transit[1] 1.116829lambda_asturias_work <- data_asturias %>%
features(mob_workplaces, features = guerrero) %>%
pull(lambda_guerrero)
lambda_asturias_work[1] 1.999927fit_model <- data_asturias_train %>%
model(
arima_at1 = ARIMA(box_cox(num_casos, lambda_asturias_casos) ~
box_cox(tmed, lambda_asturias_tmed) +
box_cox(mob_grocery_pharmacy, lambda_asturias_grocery) +
box_cox(mob_parks, lambda_asturias_parks) +
box_cox(mob_residential, lambda_asturias_resd) +
box_cox(mob_retail_recreation, lambda_asturias_retail) +
box_cox(mob_transit_stations, lambda_asturias_transit) +
box_cox(mob_workplaces, lambda_asturias_work)
),
arima_at2 = ARIMA(box_cox(num_casos, lambda_asturias_casos) ~
box_cox(tmed, lambda_asturias_tmed) +
box_cox(mob_grocery_pharmacy, lambda_asturias_grocery) +
box_cox(mob_parks, lambda_asturias_parks) +
box_cox(mob_residential, lambda_asturias_resd) +
box_cox(mob_retail_recreation, lambda_asturias_retail) +
box_cox(mob_transit_stations, lambda_asturias_transit) +
box_cox(mob_workplaces, lambda_asturias_work),
stepwise = FALSE, approx = FALSE),
Snaive = SNAIVE(box_cox(num_casos, lambda_asturias_casos))
)
# Show and report model
fit_model# A mable: 1 x 4
# Key: provincia [1]
provincia arima_at1
<chr> <model>
1 Asturias <LM w/ ARIMA(3,1,1)(2,0,0)[7] errors>
# … with 2 more variables: arima_at2 <model>, Snaive <model>glance(fit_model)# A tibble: 3 × 9
provincia .model sigma2 log_lik AIC AICc BIC ar_roots ma_roots
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <list> <list>
1 Asturias arima_at1 0.916 -812. 1653. 1653. 1714. <cpl [17]> <cpl [1]>
2 Asturias arima_at2 0.891 -806. 1634. 1635. 1683. <cpl [7]> <cpl [8]>
3 Asturias Snaive 2.53 NA NA NA NA <NULL> <NULL> fit_model %>%
pivot_longer(-provincia,
names_to = ".model",
values_to = "orders") %>%
left_join(glance(fit_model), by = c("provincia", ".model")) %>%
arrange(AICc) %>%
select(.model:BIC)# A mable: 3 x 7
# Key: .model [3]
.model orders sigma2 log_lik AIC AICc BIC
<chr> <model> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_… <LM w/ ARIMA(0,1,1)(1,0,1)[7] errors> 0.891 -806. 1634. 1635. 1683.
2 arima_… <LM w/ ARIMA(3,1,1)(2,0,0)[7] errors> 0.916 -812. 1653. 1653. 1714.
3 Snaive <SNAIVE> 2.53 NA NA NA NA # We use a Ljung-Box test >> large p-value, confirms residuals are similar / considered to white noise
fit_model %>%
select(Snaive) %>%
gg_tsresiduals()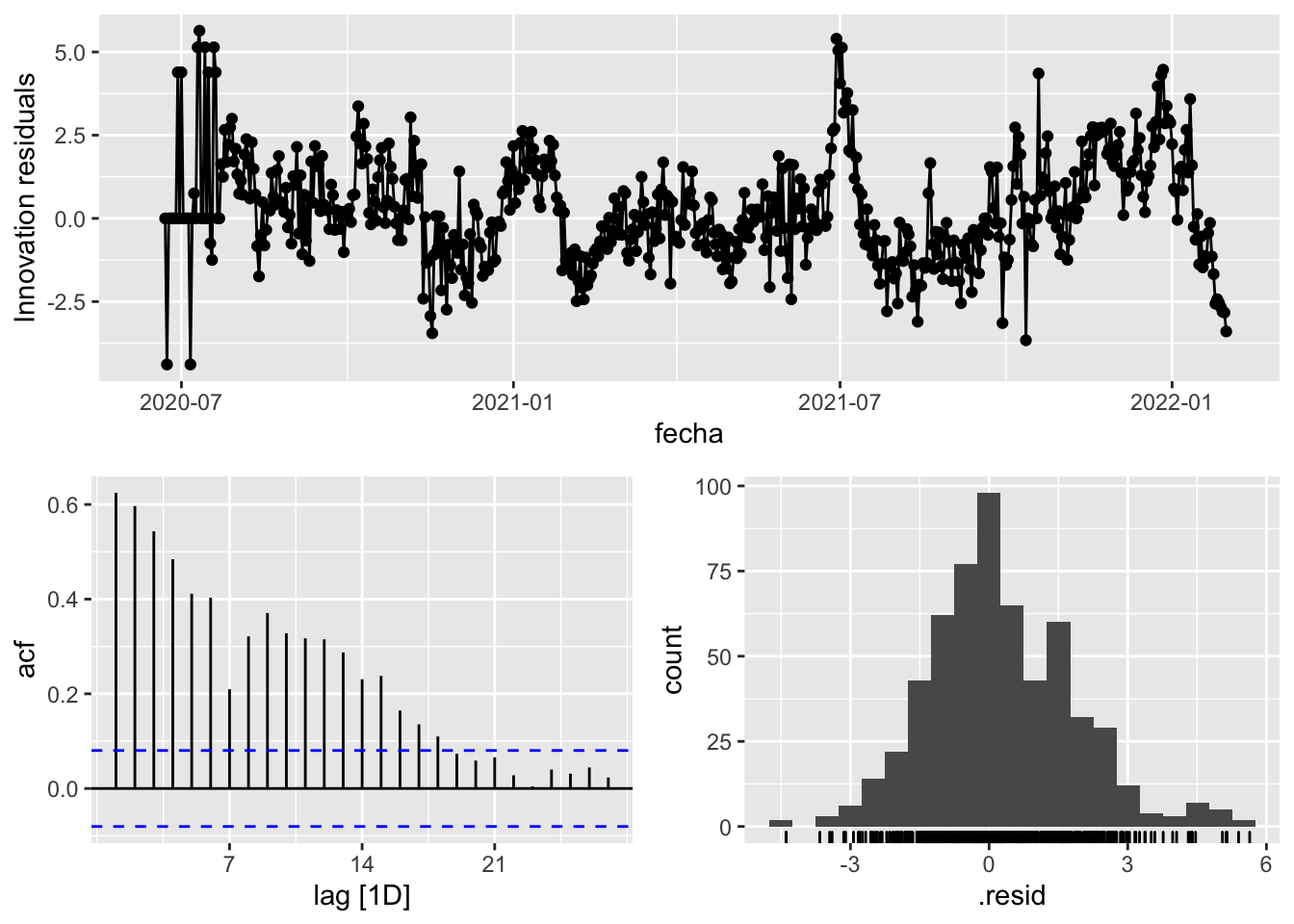
fit_model %>%
select(arima_at1) %>%
gg_tsresiduals()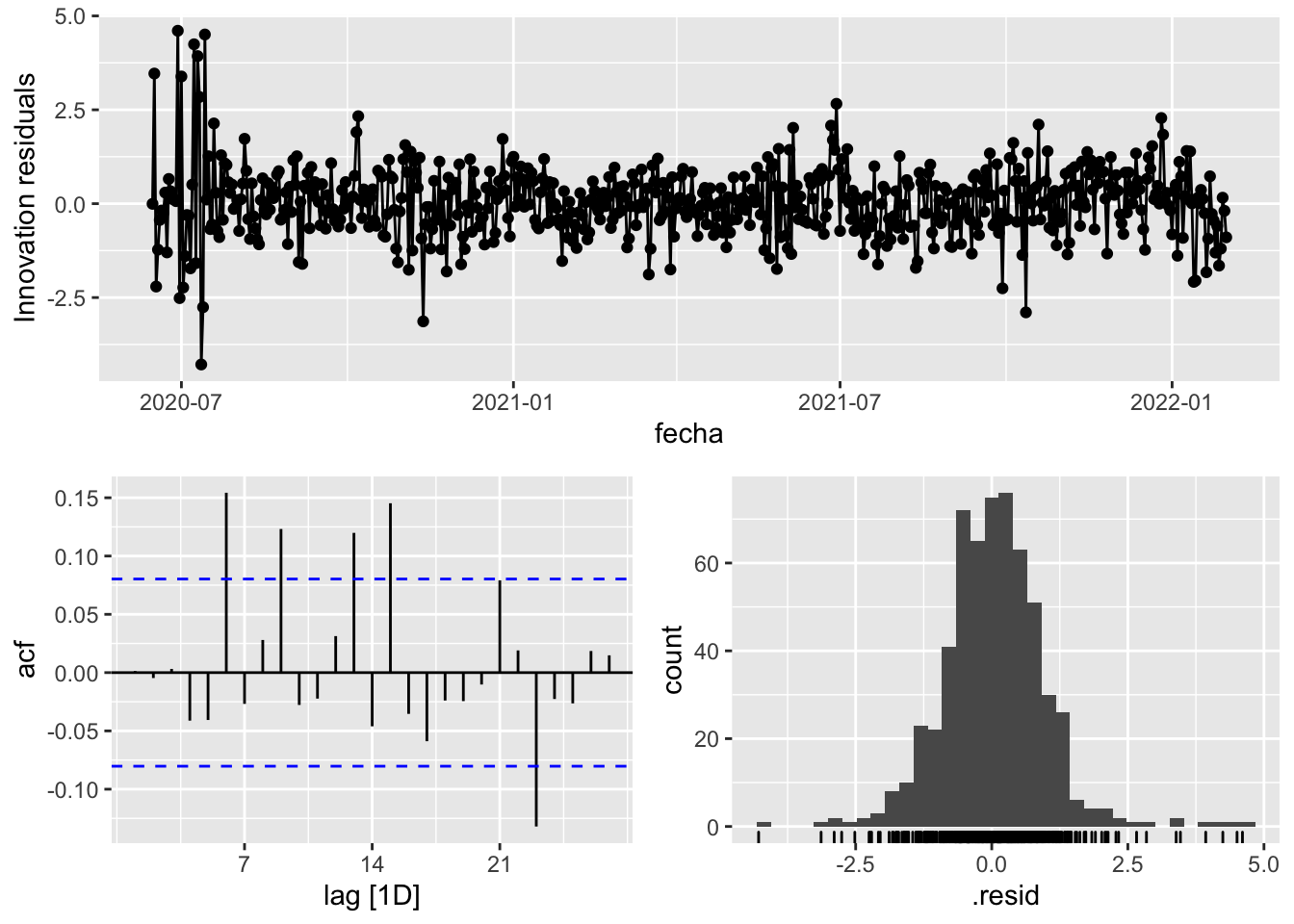
fit_model %>%
select(arima_at2) %>%
gg_tsresiduals()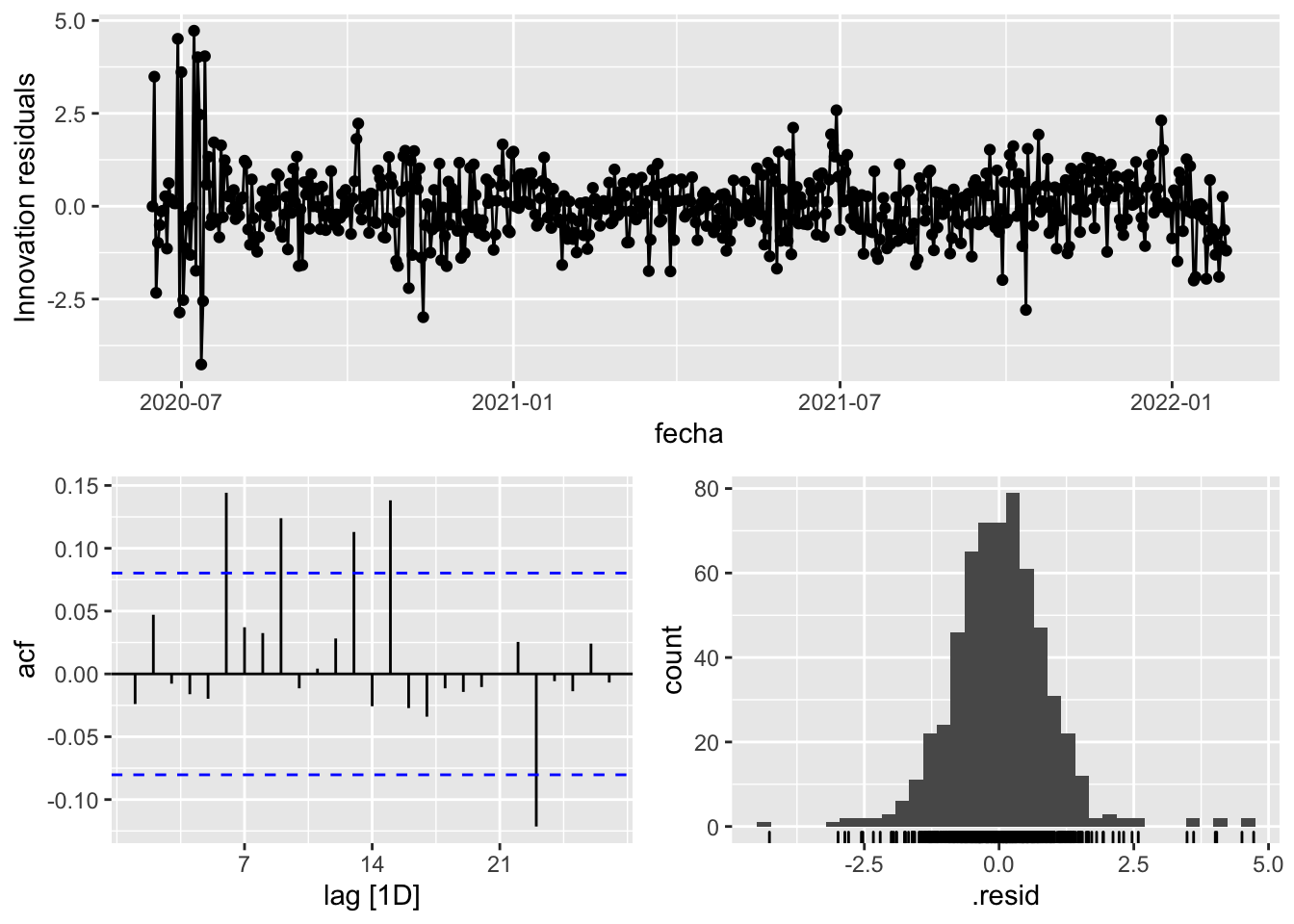
augment(fit_model) %>%
features(.innov, ljung_box, lag=7)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Asturias arima_at1 16.9 0.0184
2 Asturias arima_at2 15.5 0.0301
3 Asturias Snaive 981. 0 augment(fit_model) %>%
features(.innov, ljung_box, lag=14)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Asturias arima_at1 38.0 0.000521
2 Asturias arima_at2 34.2 0.00190
3 Asturias Snaive 1393. 0 augment(fit_model) %>%
features(.innov, ljung_box, lag=21)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Asturias arima_at1 58.5 0.0000214
2 Asturias arima_at2 47.4 0.000829
3 Asturias Snaive 1470. 0 augment(fit_model) %>%
features(.innov, ljung_box, lag=57)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Asturias arima_at1 103. 0.000188
2 Asturias arima_at2 79.8 0.0249
3 Asturias Snaive 1527. 0 # To predict future values of infections, we need to pass the model future/prediction values of the complementary variables tmed and mobility
# We are going to select those from the test dataset
data_fc_h7 <- data_asturias_test %>%
select(-num_casos) %>%
slice(1:7)
data_fc_h14 <- data_asturias_test %>%
select(-num_casos) %>%
slice(1:14)
data_fc_h21 <- data_asturias_test %>%
select(-num_casos) %>%
slice(1:21)
data_fc_h57 <- data_asturias_test %>%
select(-num_casos) %>%
slice(1:57)# Significant spikes out of 30 is still consistent with white noise
# To be sure, use a Ljung-Box test, which has a large p-value, confirming that
# the residuals are similar to white noise.
# Note that the alternative models also passed this test.
#
# Forecast
fc_h7 <- fabletools::forecast(fit_model, new_data = data_fc_h7)
fc_h14 <- fabletools::forecast(fit_model, new_data = data_fc_h14)
fc_h21 <- fabletools::forecast(fit_model, new_data = data_fc_h21)
fc_h57 <- fabletools::forecast(fit_model, new_data = data_fc_h57)7 days
# Plots
fc_h7 %>%
autoplot(
data_asturias_test %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(7))
) +
labs(y = "Nº cases", title = "Asturias - forecast h7")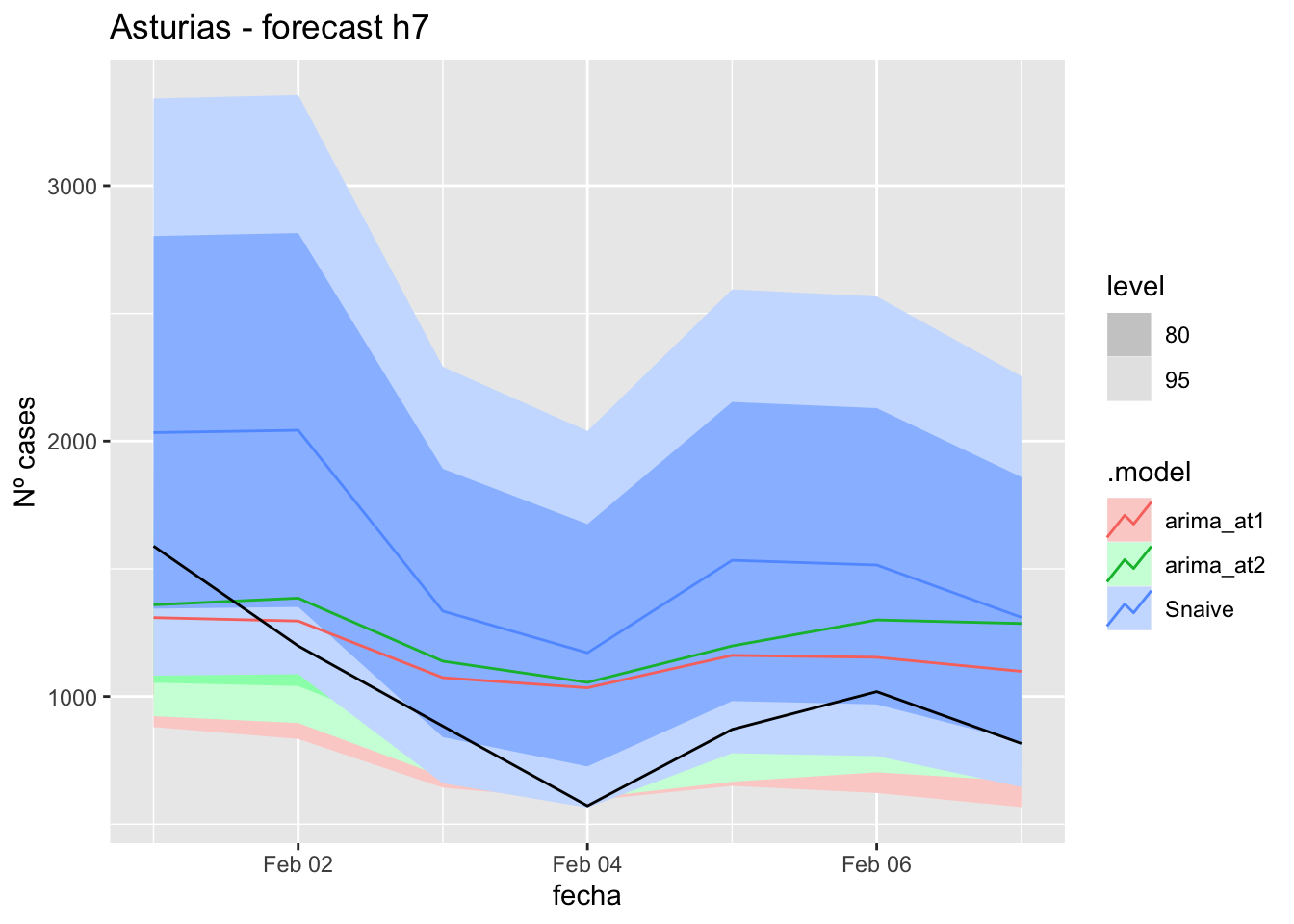
fc_h7 %>%
autoplot(
data_asturias %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(7) &
fecha > as.Date("2022-01-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Asturias - forecast h7")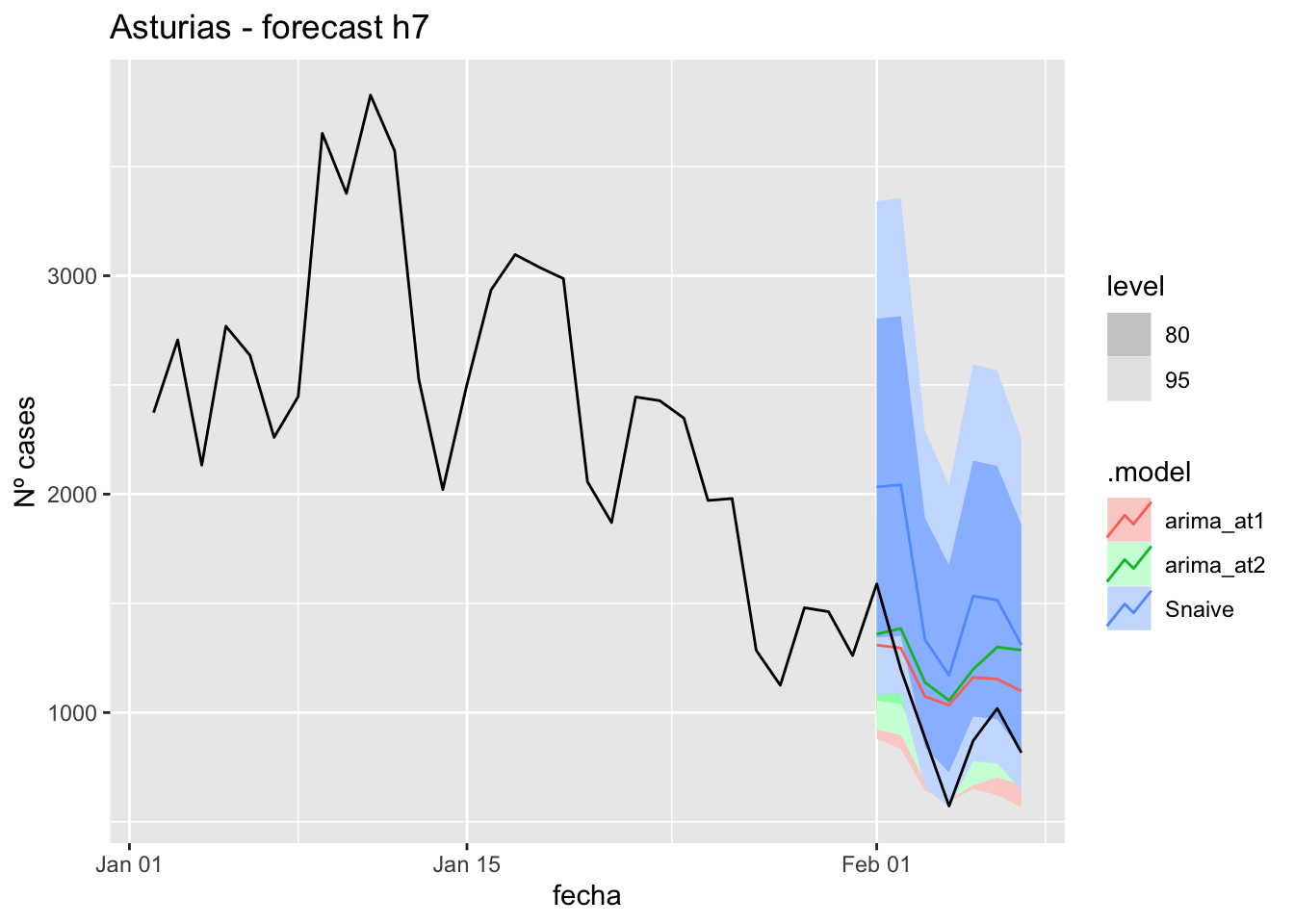
# Accuracy
fabletools::accuracy(fc_h7, data_asturias)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_at1 Asturias Test -168. 272. 248. -24.8 29.9 2.56 1.24 0.200
2 arima_at2 Asturias Test -253. 336. 319. -33.9 38.0 3.29 1.53 0.166
3 Snaive Asturias Test -570. 586. 570. -62.8 62.8 5.89 2.66 -0.55014 days
# Plots
fc_h14 %>%
autoplot(
data_asturias_test %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(14))
) +
labs(y = "Nº cases", title = "Asturias - forecast h14")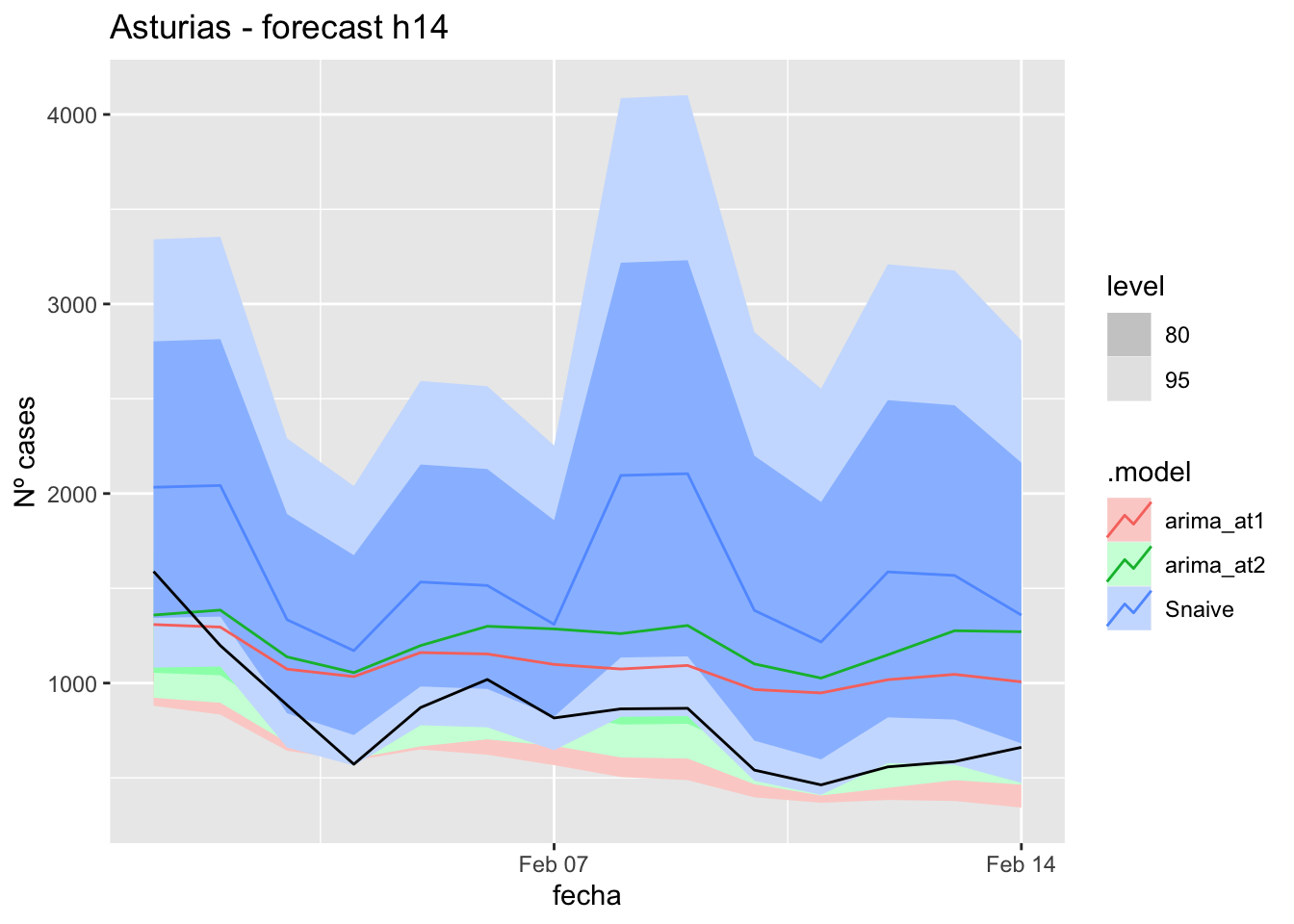
fc_h14 %>%
autoplot(
data_asturias %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(14) &
fecha > as.Date("2022-01-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Asturias - forecast h14")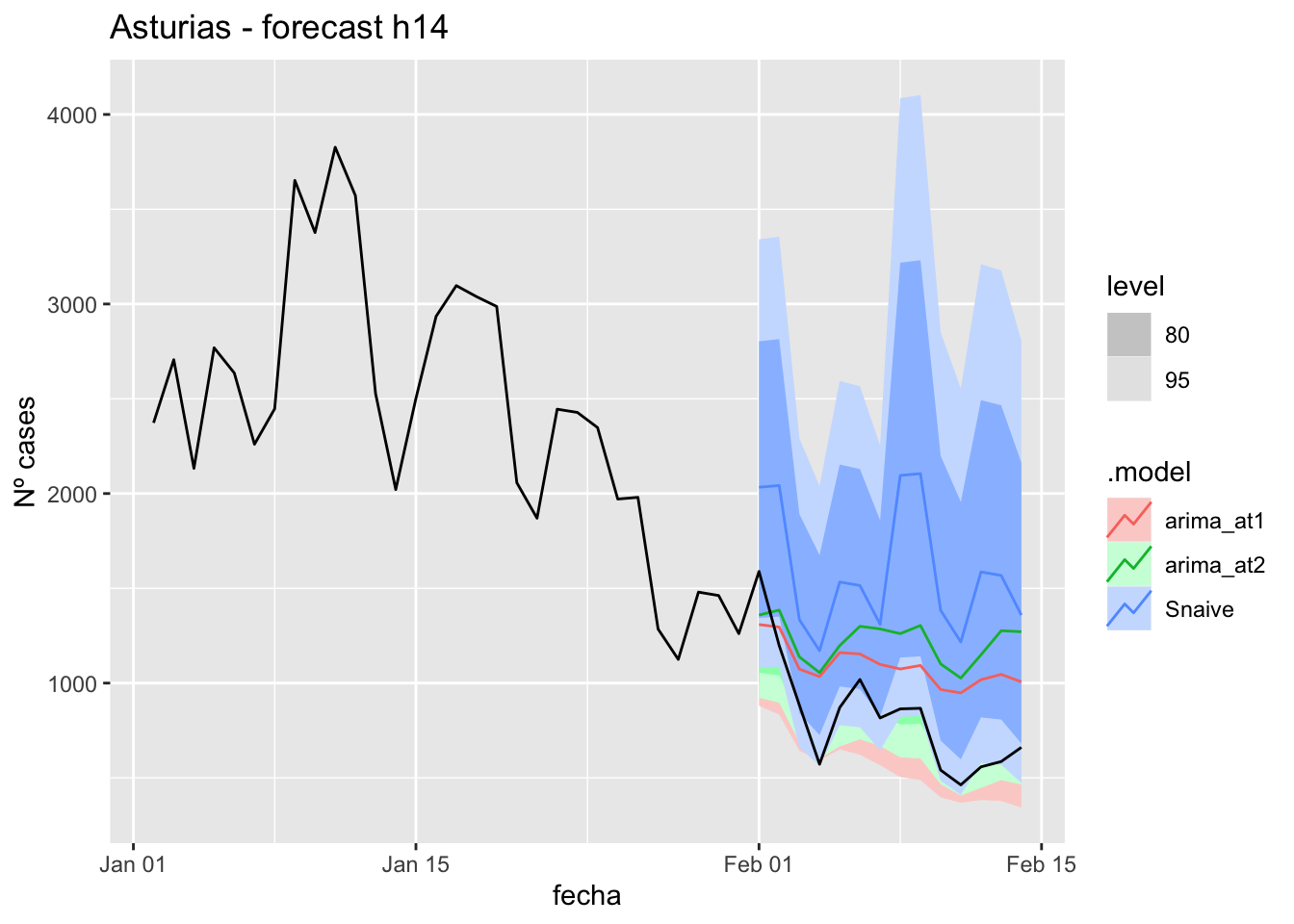
# Accuracy
fabletools::accuracy(fc_h14, data_asturias)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_at1 Asturias Test -271. 335. 311. -44.3 46.9 3.21 1.52 0.387
2 arima_at2 Asturias Test -402. 461. 434. -62.5 64.5 4.49 2.09 0.457
3 Snaive Asturias Test -769. 812. 769. -107. 107. 7.94 3.69 0.29921 days
# Plots
fc_h21 %>%
autoplot(
data_asturias_test %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(21))
) +
labs(y = "Nº cases", title = "Asturias - forecast h21")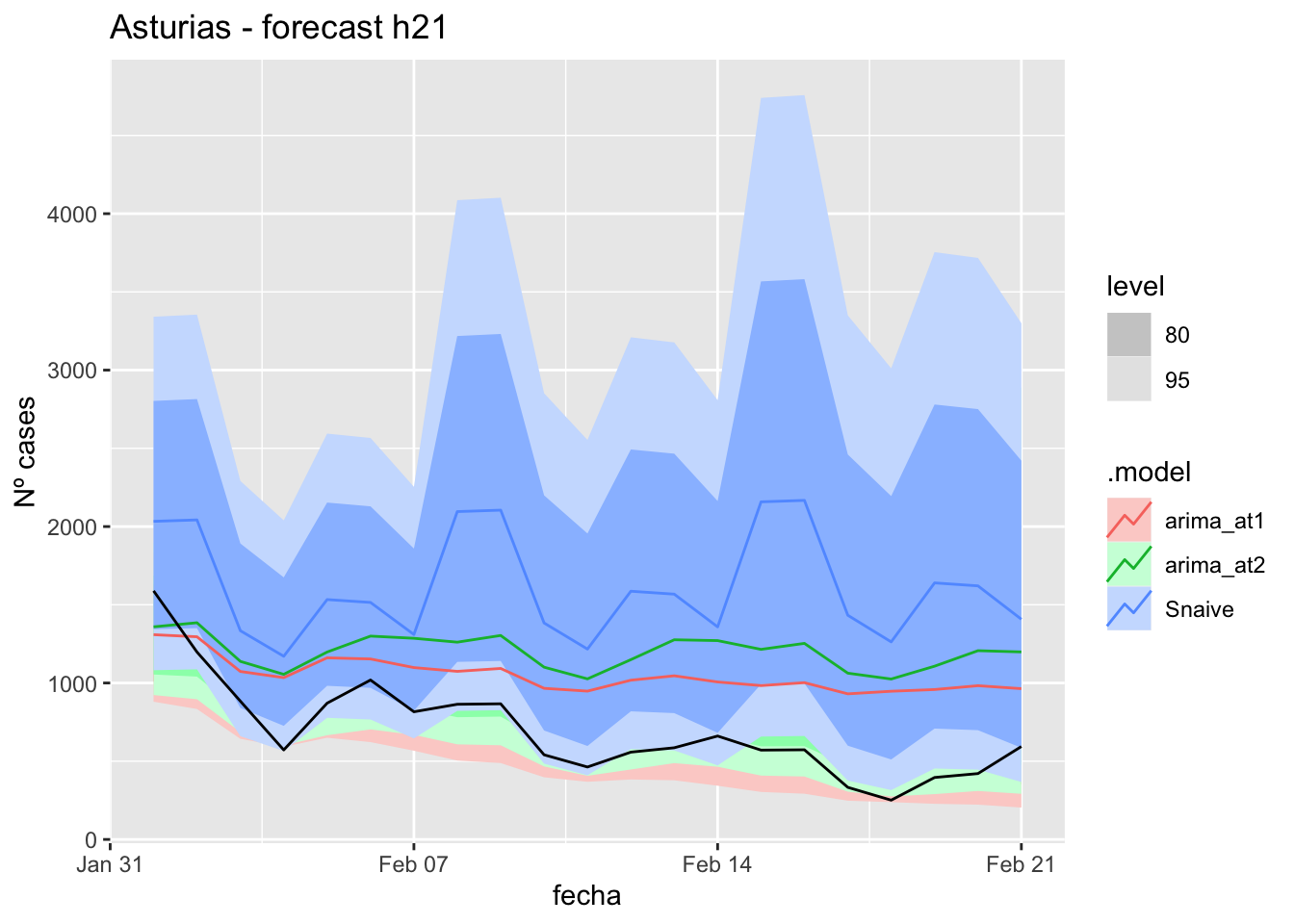
fc_h21 %>%
autoplot(
data_asturias %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(21) &
fecha > as.Date("2022-01-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Asturias - forecast h21")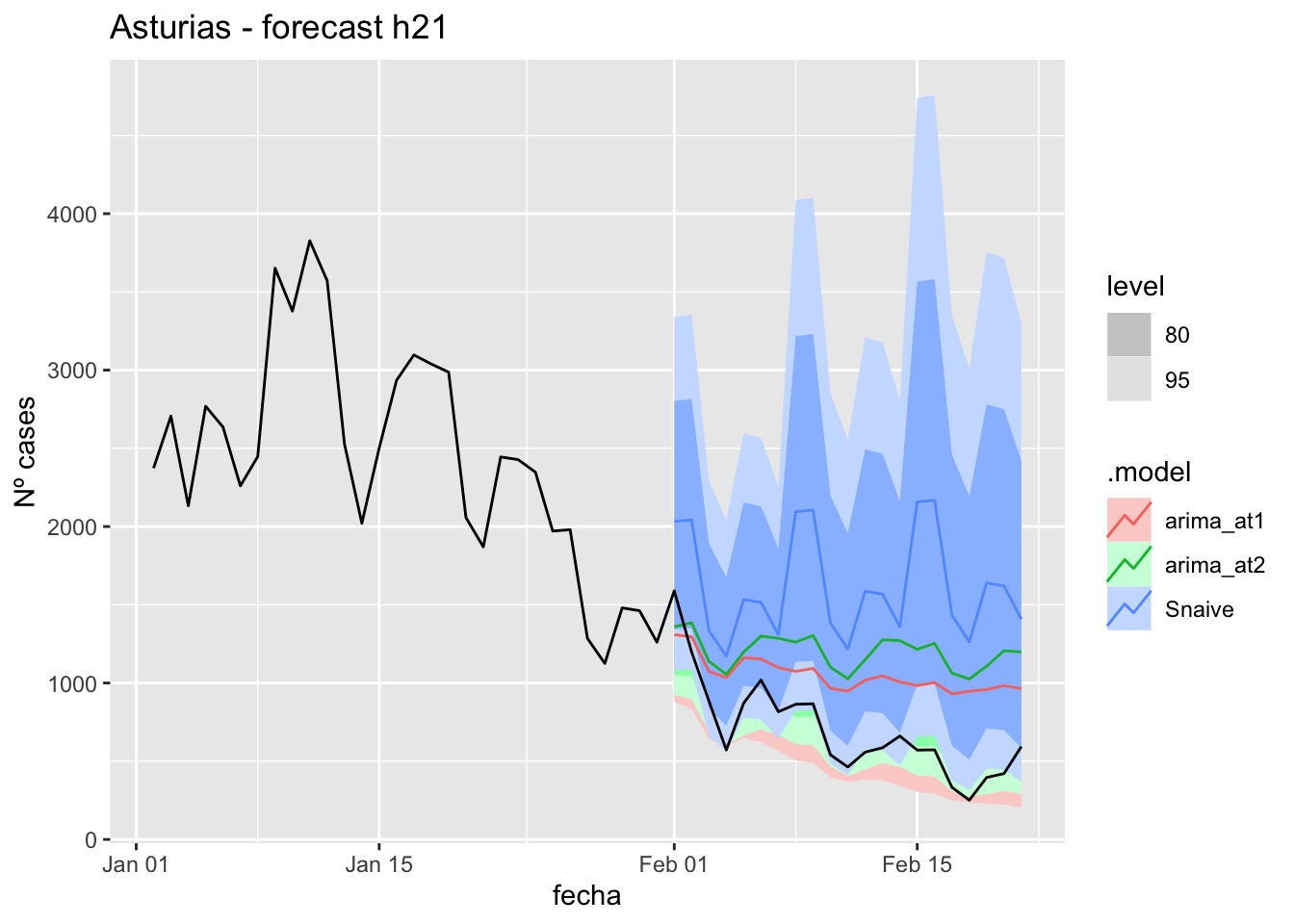
# Accuracy
fabletools::accuracy(fc_h21, data_asturias)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_at1 Asturias Test -353. 410. 380. -74.4 76.1 3.93 1.87 0.530
2 arima_at2 Asturias Test -502. 555. 524. -100. 101. 5.42 2.52 0.602
3 Snaive Asturias Test -920. 980. 920. -168. 168. 9.50 4.46 0.43757 days
# Plots
fc_h57 %>%
autoplot(
data_asturias_test %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(57))
) +
labs(y = "Nº cases", title = "Asturias - forecast h57")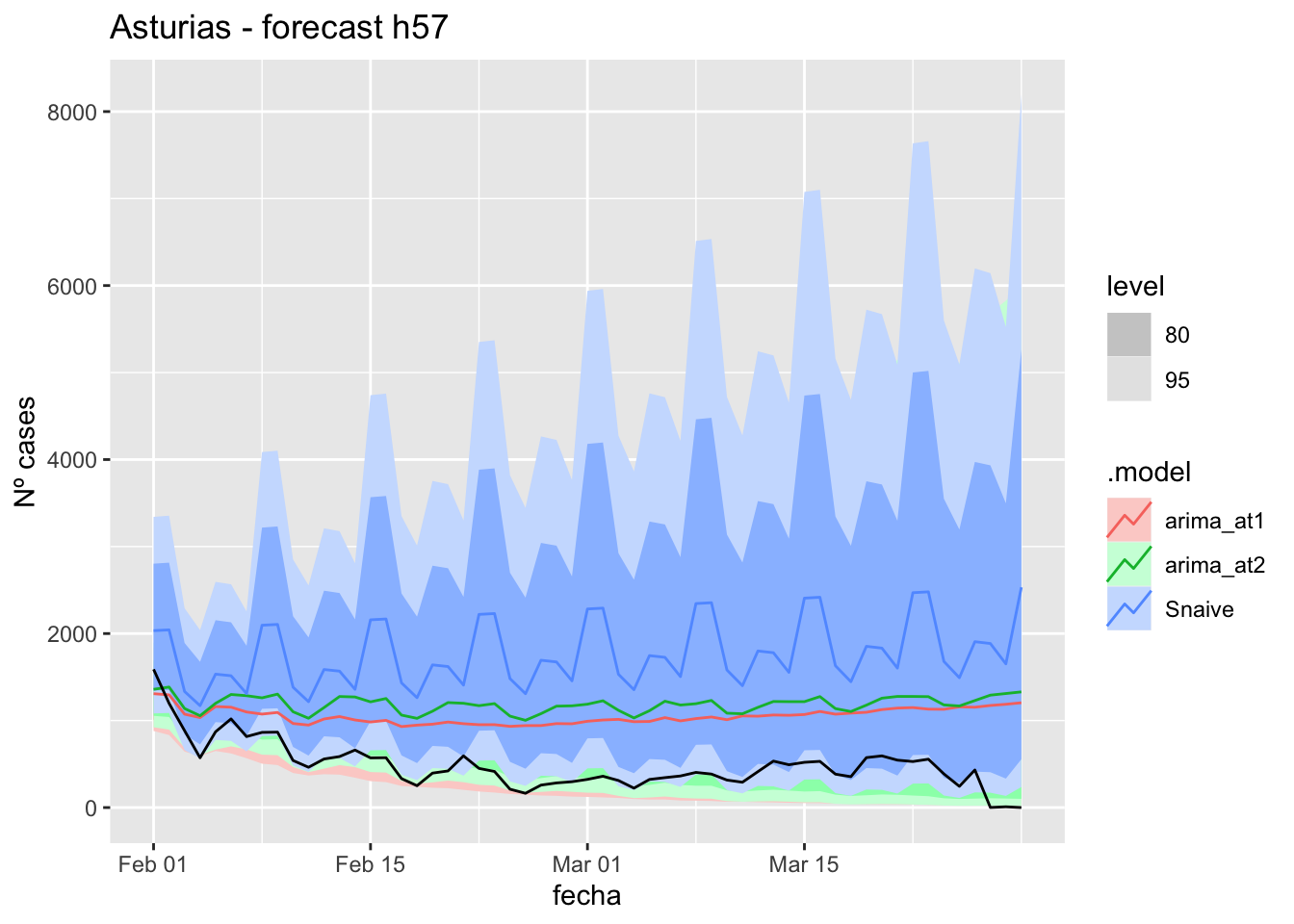
fc_h57 %>%
autoplot(
data_asturias %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(57) &
fecha > as.Date("2022-01-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Asturias - forecast h57")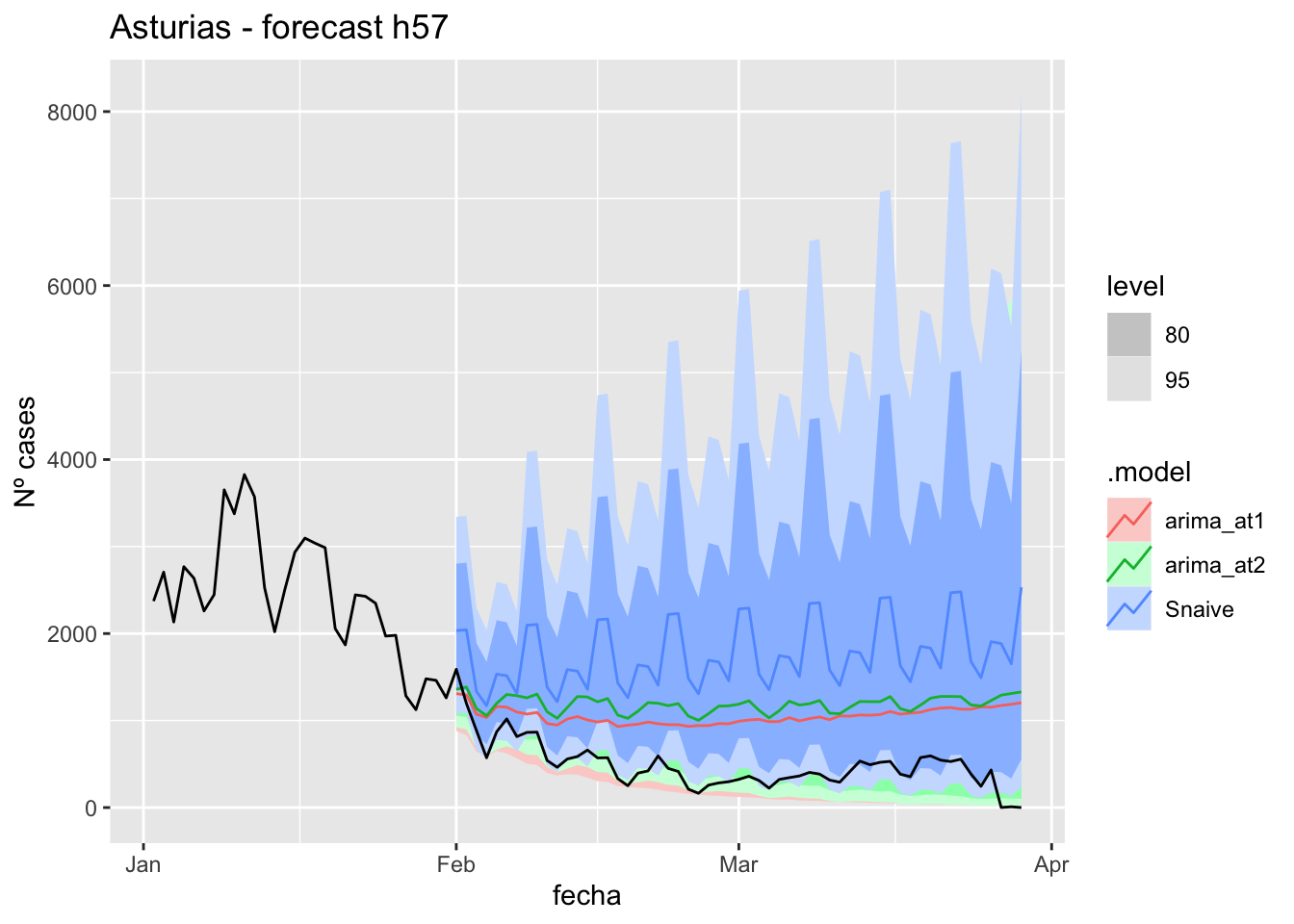
# Accuracy
fabletools::accuracy(fc_h57, data_asturias)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_at1 Asturias Test -571. 623. 581. -Inf Inf 6.00 2.83 0.706
2 arima_at2 Asturias Test -707. 749. 715. -Inf Inf 7.39 3.41 0.710
3 Snaive Asturias Test -1282. 1359. 1282. -Inf Inf 13.3 6.18 0.471Barcelona
data_Barcelona <- covid_data %>%
filter(provincia == "Barcelona") %>%
filter(fecha > as.Date("2020-06-14", format = "%Y-%m-%d")) %>%
select(provincia, fecha, num_casos, tmed, mob_grocery_pharmacy, mob_parks, mob_residential, mob_retail_recreation, mob_transit_stations, mob_workplaces) %>%
drop_na() %>%
as_tsibble(key = provincia, index = fecha)
data_Barcelona# A tsibble: 653 x 10 [1D]
# Key: provincia [1]
provincia fecha num_casos tmed mob_grocery_pharmacy mob_parks
<chr> <date> <dbl> <dbl> <dbl> <dbl>
1 Barcelona 2020-06-15 62 21.2 -9 17
2 Barcelona 2020-06-16 66 20.8 -10 -16
3 Barcelona 2020-06-17 70 20.3 -4 14
4 Barcelona 2020-06-18 68 18.8 -8 -6
5 Barcelona 2020-06-19 65 20.4 -5 10
6 Barcelona 2020-06-20 53 21.8 -12 15
7 Barcelona 2020-06-21 38 22.8 -18 18
8 Barcelona 2020-06-22 69 23.8 5 24
9 Barcelona 2020-06-23 95 23.4 19 28
10 Barcelona 2020-06-24 44 23.6 -71 28
# … with 643 more rows, and 4 more variables: mob_residential <dbl>,
# mob_retail_recreation <dbl>, mob_transit_stations <dbl>,
# mob_workplaces <dbl>data_Barcelona_train <- data_Barcelona %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d"))
summary(data_Barcelona_train$fecha) Min. 1st Qu. Median Mean 3rd Qu. Max.
"2020-06-15" "2020-11-10" "2021-04-08" "2021-04-08" "2021-09-04" "2022-01-31" data_Barcelona_test <- data_Barcelona %>%
filter(fecha > as.Date("2022-01-31", format = "%Y-%m-%d"))
summary(data_Barcelona_test$fecha) Min. 1st Qu. Median Mean 3rd Qu. Max.
"2022-02-01" "2022-02-15" "2022-03-01" "2022-03-01" "2022-03-15" "2022-03-29" # Lamba for num_casos
lambda_Barcelona_casos <- data_asturias %>%
features(num_casos, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Barcelona_casos[1] 0.2278229# Lamba for average temp
lambda_Barcelona_tmed <- data_asturias %>%
features(tmed, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Barcelona_tmed[1] 1.056074# Lamba for mobility
lambda_Barcelona_grocery <- data_asturias %>%
features(mob_grocery_pharmacy, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Barcelona_grocery[1] 1.377952lambda_Barcelona_parks <- data_asturias %>%
features(mob_parks, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Barcelona_parks[1] 0.7188469lambda_Barcelona_resd <- data_asturias %>%
features(mob_residential, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Barcelona_resd[1] 1.000064lambda_Barcelona_retail <- data_asturias %>%
features(mob_retail_recreation, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Barcelona_retail[1] 1.058286lambda_Barcelona_transit <- data_asturias %>%
features(mob_transit_stations, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Barcelona_transit[1] 1.116829lambda_Barcelona_work <- data_asturias %>%
features(mob_workplaces, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Barcelona_work[1] 1.999927fit_model <- data_Barcelona_train %>%
model(
arima_at1 = ARIMA(box_cox(num_casos, lambda_Barcelona_casos) ~
box_cox(tmed, lambda_Barcelona_tmed) +
box_cox(mob_grocery_pharmacy, lambda_Barcelona_grocery) +
box_cox(mob_parks, lambda_Barcelona_parks) +
box_cox(mob_residential, lambda_Barcelona_resd) +
box_cox(mob_retail_recreation, lambda_Barcelona_retail) +
box_cox(mob_transit_stations, lambda_Barcelona_transit) +
box_cox(mob_workplaces, lambda_Barcelona_work)
),
arima_at2 = ARIMA(box_cox(num_casos, lambda_Barcelona_casos) ~
box_cox(tmed, lambda_Barcelona_tmed) +
box_cox(mob_grocery_pharmacy, lambda_Barcelona_grocery) +
box_cox(mob_parks, lambda_Barcelona_parks) +
box_cox(mob_residential, lambda_Barcelona_resd) +
box_cox(mob_retail_recreation, lambda_Barcelona_retail) +
box_cox(mob_transit_stations, lambda_Barcelona_transit) +
box_cox(mob_workplaces, lambda_Barcelona_work),
stepwise = FALSE, approx = FALSE),
Snaive = SNAIVE(box_cox(num_casos, lambda_Barcelona_casos))
)
# Show and report model
fit_model# A mable: 1 x 4
# Key: provincia [1]
provincia arima_at1
<chr> <model>
1 Barcelona <LM w/ ARIMA(1,1,0)(1,0,0)[7] errors>
# … with 2 more variables: arima_at2 <model>, Snaive <model>glance(fit_model)# A tibble: 3 × 9
provincia .model sigma2 log_lik AIC AICc BIC ar_roots ma_roots
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <list> <list>
1 Barcelona arima_at1 0.909 -816. 1652. 1653. 1696. <cpl [8]> <cpl [0]>
2 Barcelona arima_at2 0.815 -783. 1591. 1592. 1648. <cpl [15]> <cpl [2]>
3 Barcelona Snaive 4.60 NA NA NA NA <NULL> <NULL> fit_model %>%
pivot_longer(-provincia,
names_to = ".model",
values_to = "orders") %>%
left_join(glance(fit_model), by = c("provincia", ".model")) %>%
arrange(AICc) %>%
select(.model:BIC)# A mable: 3 x 7
# Key: .model [3]
.model orders sigma2 log_lik AIC AICc BIC
<chr> <model> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_… <LM w/ ARIMA(1,1,2)(2,0,0)[7] errors> 0.815 -783. 1591. 1592. 1648.
2 arima_… <LM w/ ARIMA(1,1,0)(1,0,0)[7] errors> 0.909 -816. 1652. 1653. 1696.
3 Snaive <SNAIVE> 4.60 NA NA NA NA # We use a Ljung-Box test >> large p-value, confirms residuals are similar / considered to white noise
fit_model %>%
select(Snaive) %>%
gg_tsresiduals()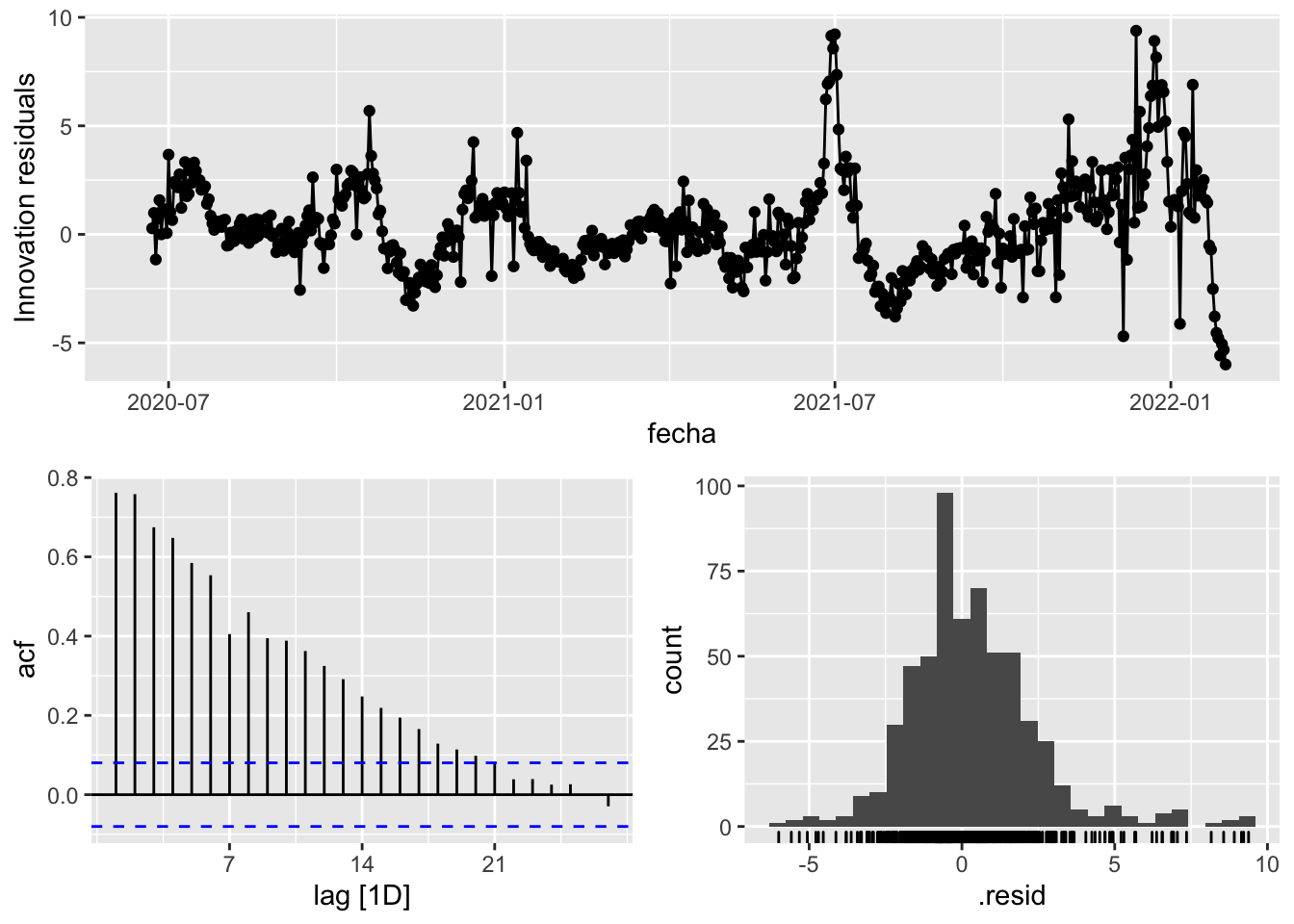
fit_model %>%
select(arima_at1) %>%
gg_tsresiduals()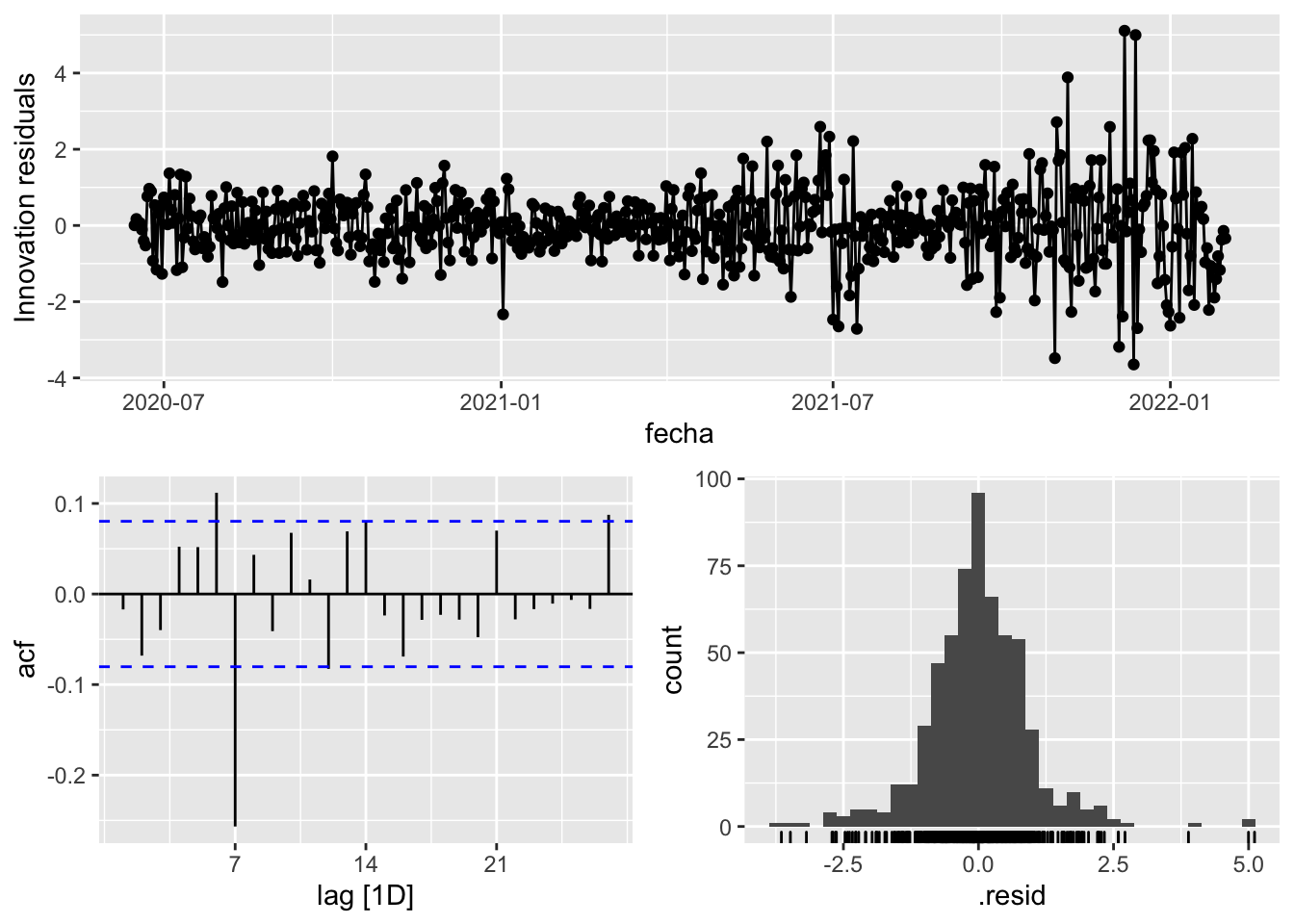
fit_model %>%
select(arima_at2) %>%
gg_tsresiduals()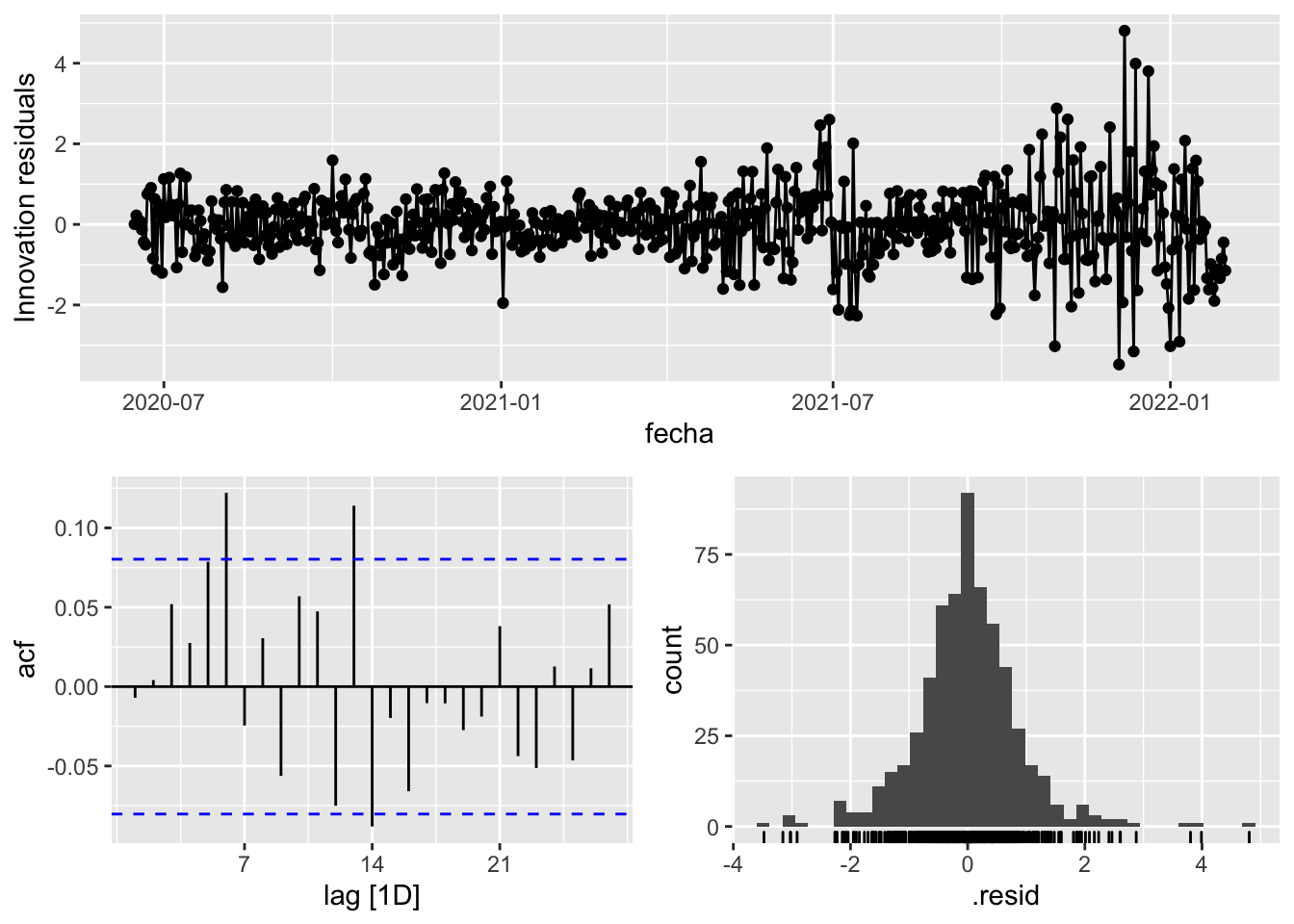
augment(fit_model) %>%
features(.innov, ljung_box, lag=7)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Barcelona arima_at1 54.6 0.00000000179
2 Barcelona arima_at2 15.2 0.0332
3 Barcelona Snaive 1689. 0 augment(fit_model) %>%
features(.innov, ljung_box, lag=14)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Barcelona arima_at1 70.7 0.00000000145
2 Barcelona arima_at2 37.2 0.000690
3 Barcelona Snaive 2232. 0 augment(fit_model) %>%
features(.innov, ljung_box, lag=21)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Barcelona arima_at1 79.7 0.00000000897
2 Barcelona arima_at2 41.8 0.00443
3 Barcelona Snaive 2328. 0 augment(fit_model) %>%
features(.innov, ljung_box, lag=57)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Barcelona arima_at1 150. 3.08e-10
2 Barcelona arima_at2 99.5 4.27e- 4
3 Barcelona Snaive 2625. 0 # To predict future values of infections, we need to pass the model future/prediction values of the complementary variables tmed and mobility
# We are going to select those from the test dataset
data_fc_h7 <- data_Barcelona_test %>%
select(-num_casos) %>%
slice(1:7)
data_fc_h14 <- data_Barcelona_test %>%
select(-num_casos) %>%
slice(1:14)
data_fc_h21 <- data_Barcelona_test %>%
select(-num_casos) %>%
slice(1:21)
data_fc_h57 <- data_Barcelona_test %>%
select(-num_casos) %>%
slice(1:57)# Significant spikes out of 30 is still consistent with white noise
# To be sure, use a Ljung-Box test, which has a large p-value, confirming that
# the residuals are similar to white noise.
# Note that the alternative models also passed this test.
#
# Forecast
fc_h7 <- fabletools::forecast(fit_model, new_data = data_fc_h7)
fc_h14 <- fabletools::forecast(fit_model, new_data = data_fc_h14)
fc_h21 <- fabletools::forecast(fit_model, new_data = data_fc_h21)
fc_h57 <- fabletools::forecast(fit_model, new_data = data_fc_h57)7 days
# Plots
fc_h7 %>%
autoplot(
data_Barcelona_test %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(7))
) +
labs(y = "Nº cases", title = "Barcelona - forecast h7")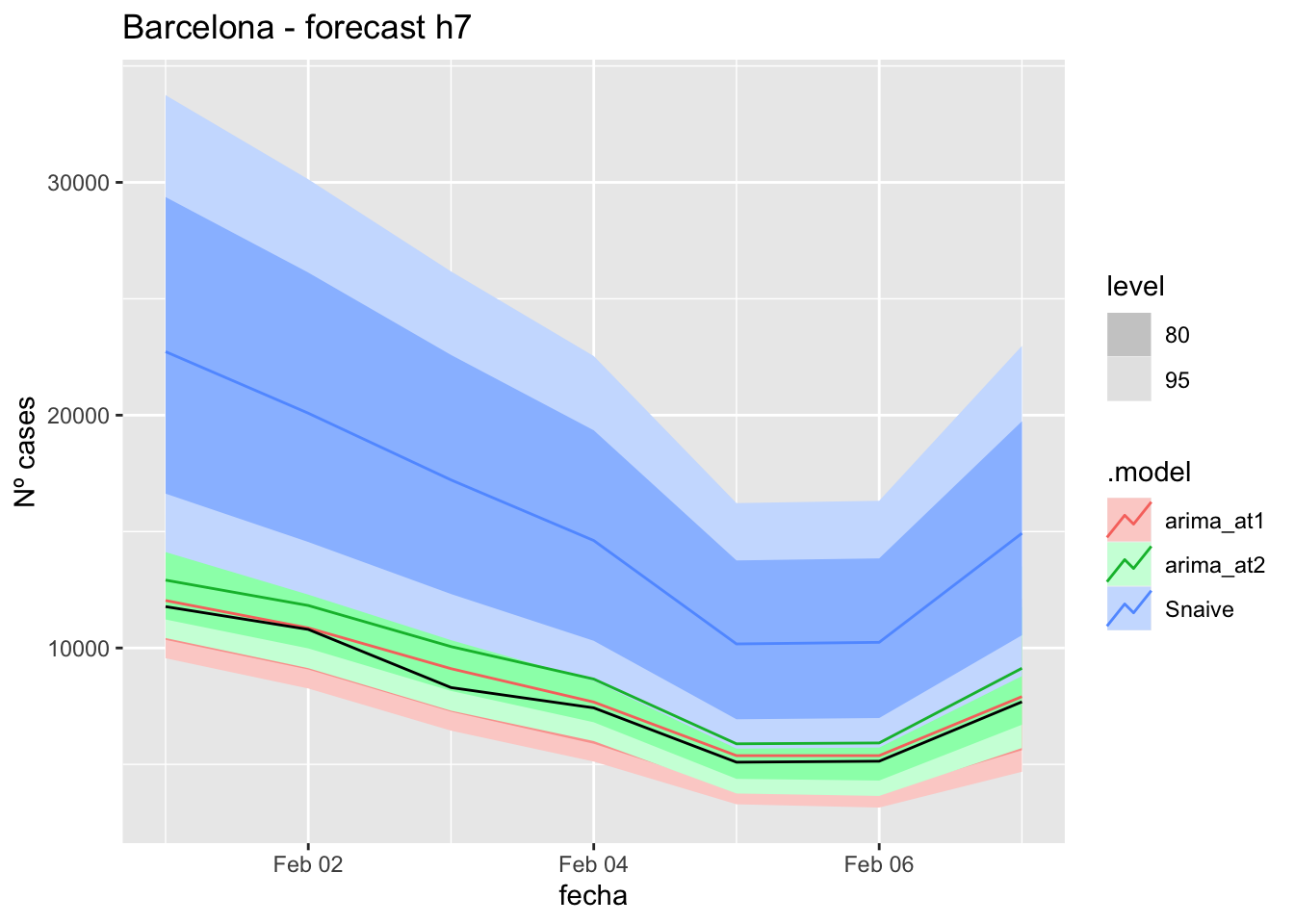
fc_h7 %>%
autoplot(
data_Barcelona %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(7) &
fecha > as.Date("2022-01-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Barcelona - forecast h7")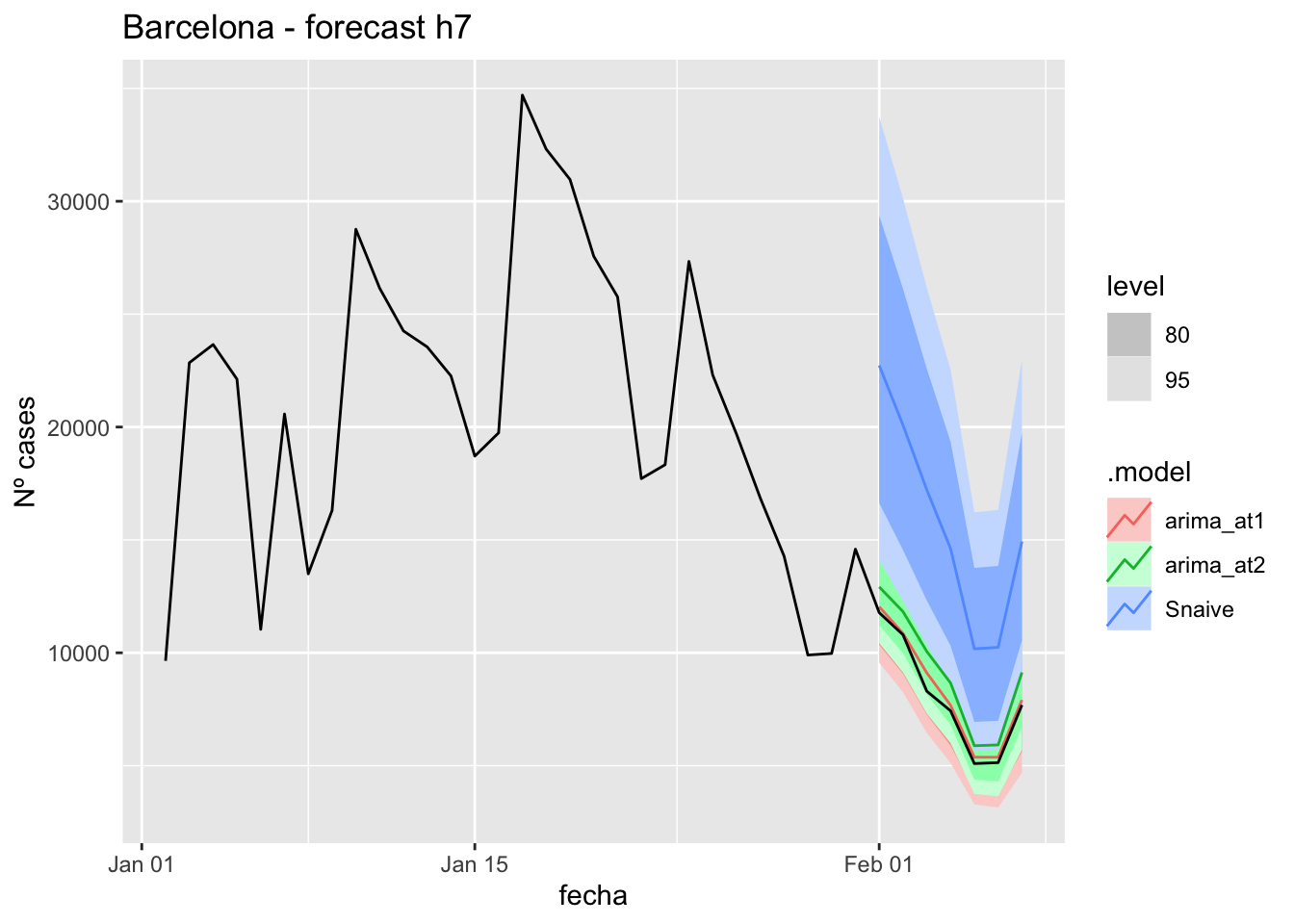
# Accuracy
fabletools::accuracy(fc_h7, data_Barcelona)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_at1 Barcelona Test -304. 374. 304. -4.15 4.15 0.382 0.191 -0.400
2 arima_at2 Barcelona Test -1168. 1213. 1168. -15.2 15.2 1.47 0.619 -0.0336
3 Snaive Barcelona Test -7681. 7943. 7681. -96.6 96.6 9.65 4.06 0.550 14 days
# Plots
fc_h14 %>%
autoplot(
data_Barcelona_test %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(14))
) +
labs(y = "Nº cases", title = "Barcelona - forecast h14")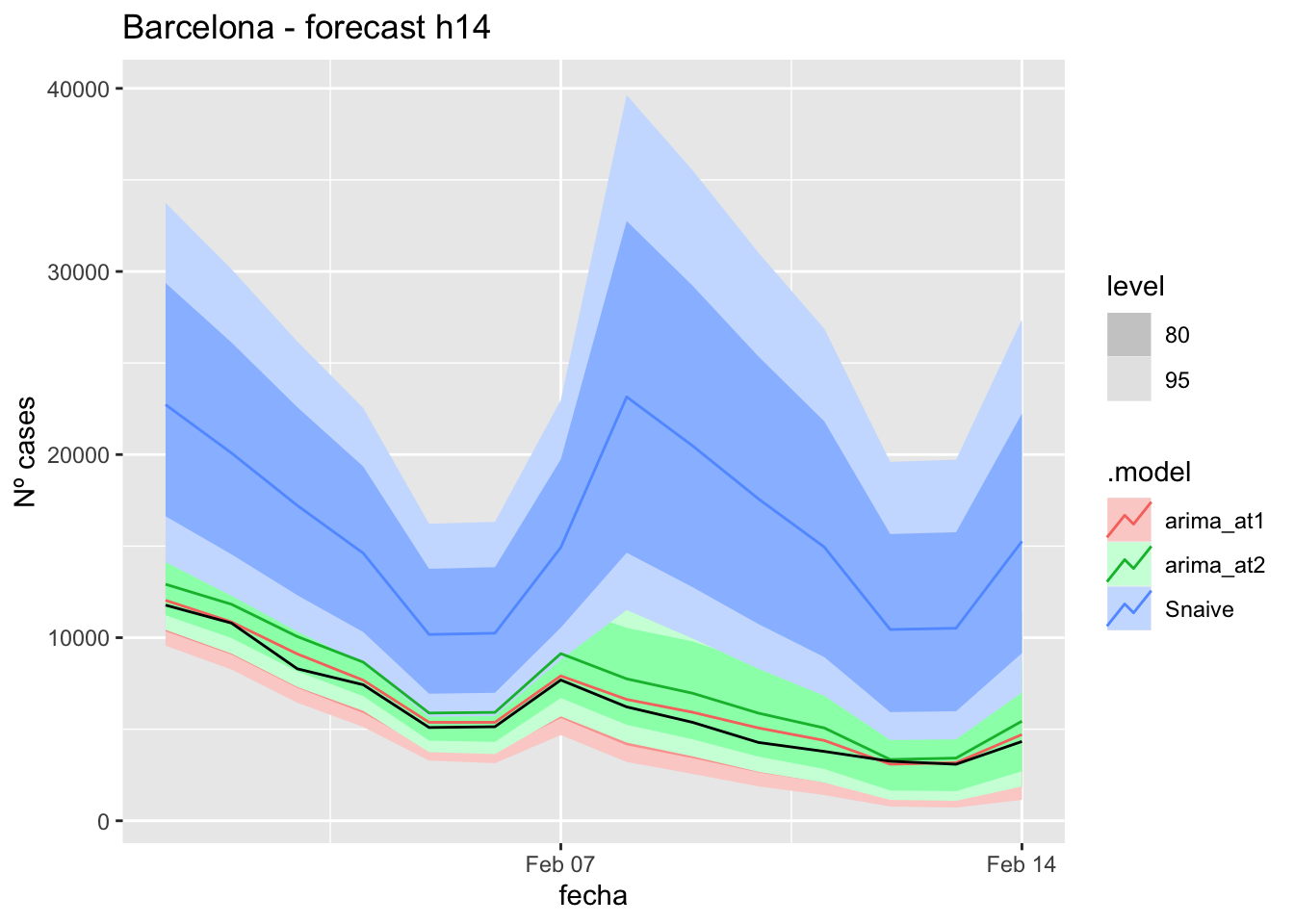
fc_h14 %>%
autoplot(
data_Barcelona %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(14) &
fecha > as.Date("2022-01-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Barcelona - forecast h14")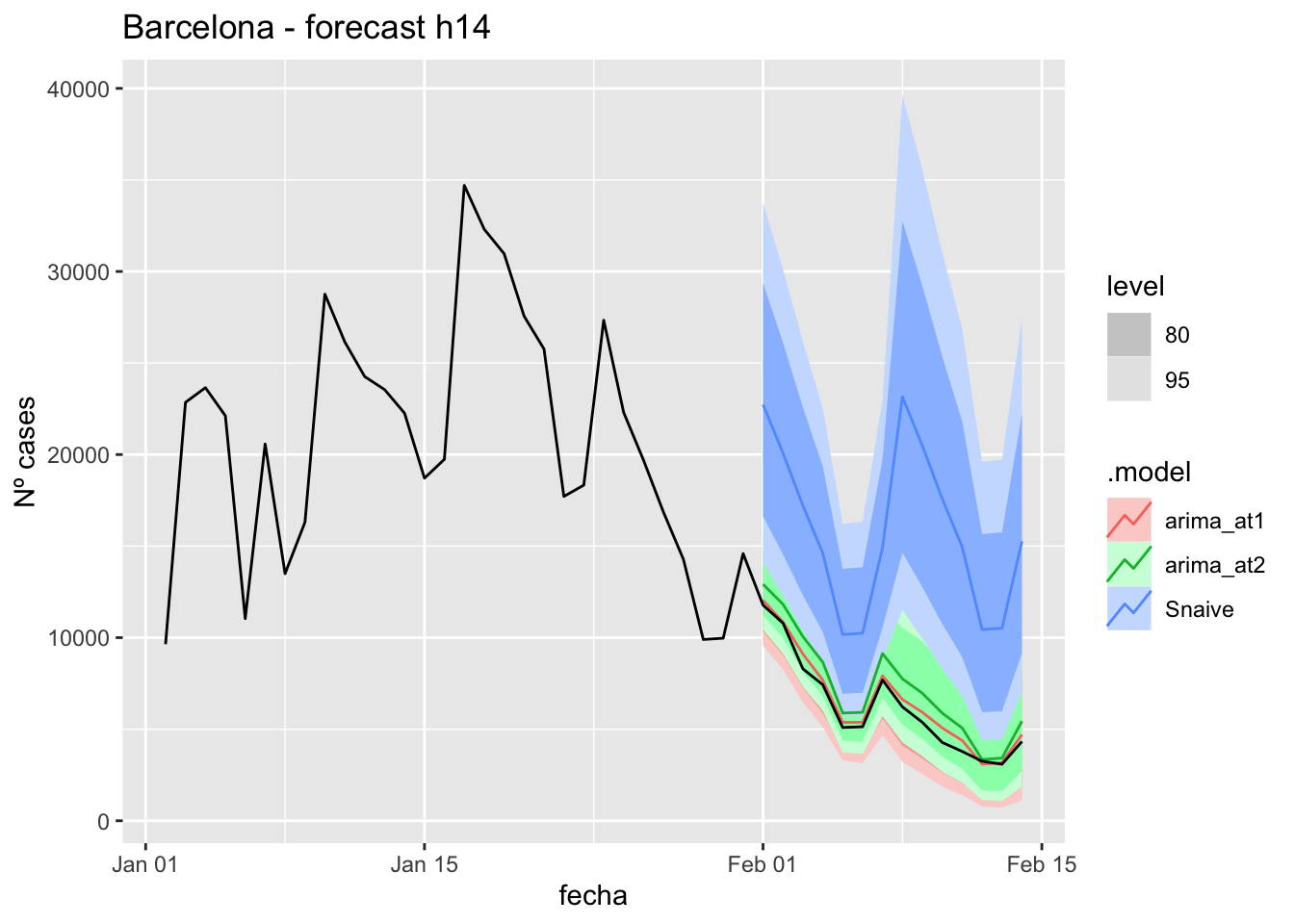
# Accuracy
fabletools::accuracy(fc_h14, data_Barcelona)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_… Barcelona Test -342. 432. 365. -6.19 6.90 0.458 0.221 0.0799
2 arima_… Barcelona Test -1123. 1217. 1123. -19.4 19.4 1.41 0.621 0.408
3 Snaive Barcelona Test -9702. 10297. 9702. -182. 182. 12.2 5.26 0.547 21 days
# Plots
fc_h21 %>%
autoplot(
data_Barcelona_test %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(21))
) +
labs(y = "Nº cases", title = "Barcelona - forecast h21")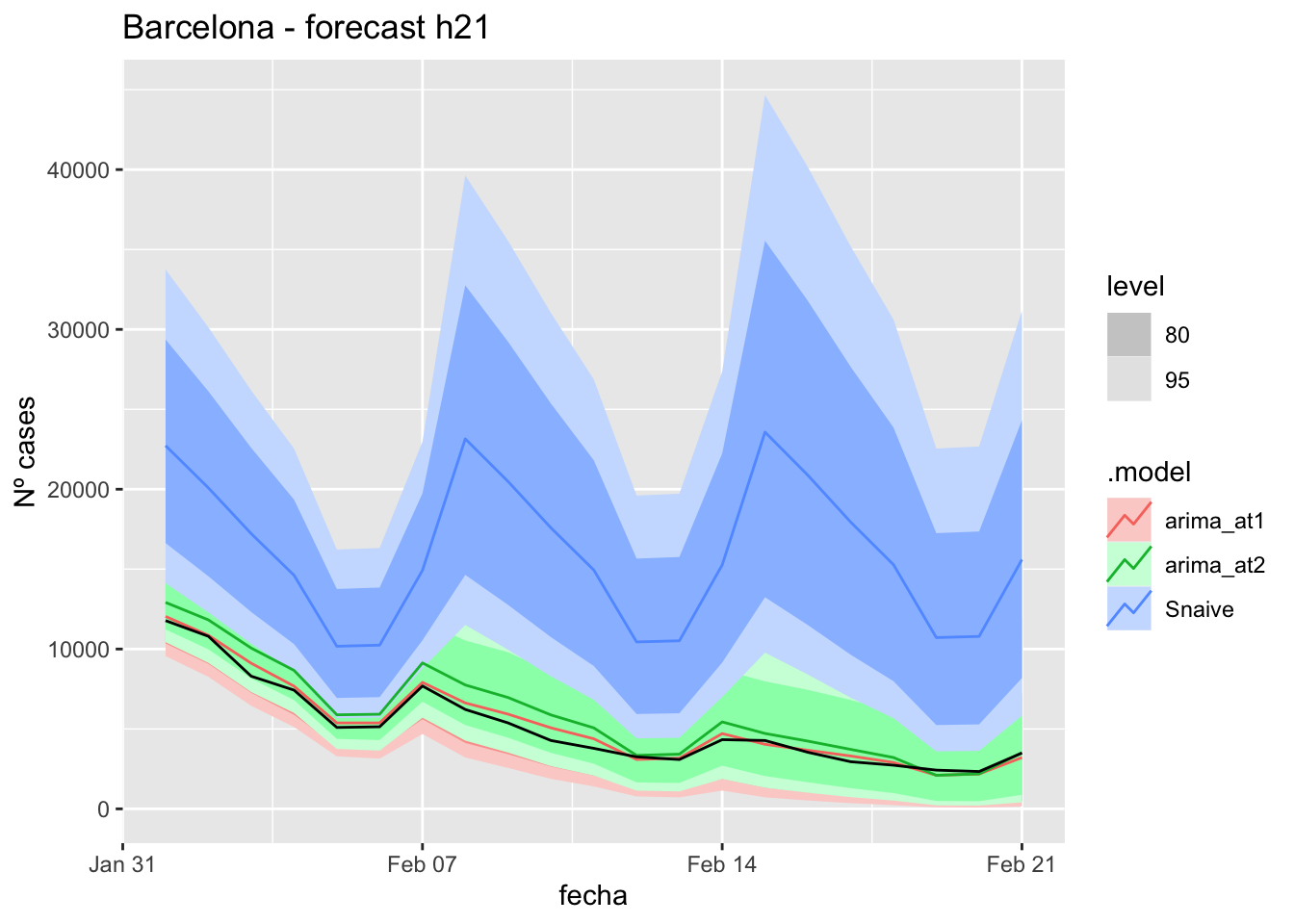
fc_h21 %>%
autoplot(
data_Barcelona %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(21) &
fecha > as.Date("2022-01-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Barcelona - forecast h21")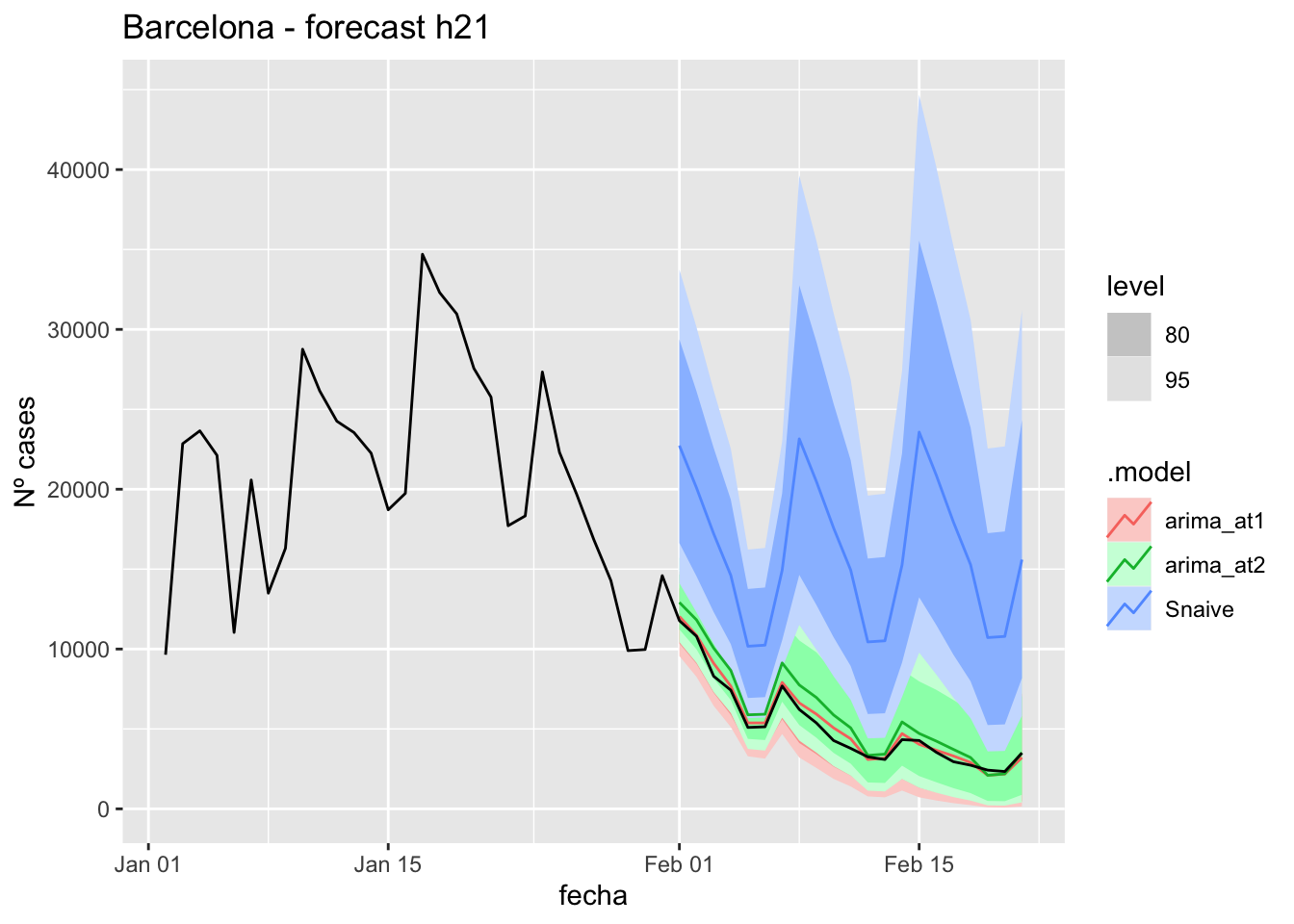
# Accuracy
fabletools::accuracy(fc_h21, data_Barcelona)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima… Barcelona Test -211. 381. 322. -3.57 7.25 0.404 0.195 0.308
2 arima… Barcelona Test -841. 1032. 886. -15.5 17.4 1.11 0.527 0.620
3 Snaive Barcelona Test -10898. 11601. 10898. -262. 262. 13.7 5.92 0.60057 days
# Plots
fc_h57 %>%
autoplot(
data_Barcelona_test %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(57))
) +
labs(y = "Nº cases", title = "Barcelona - forecast h57")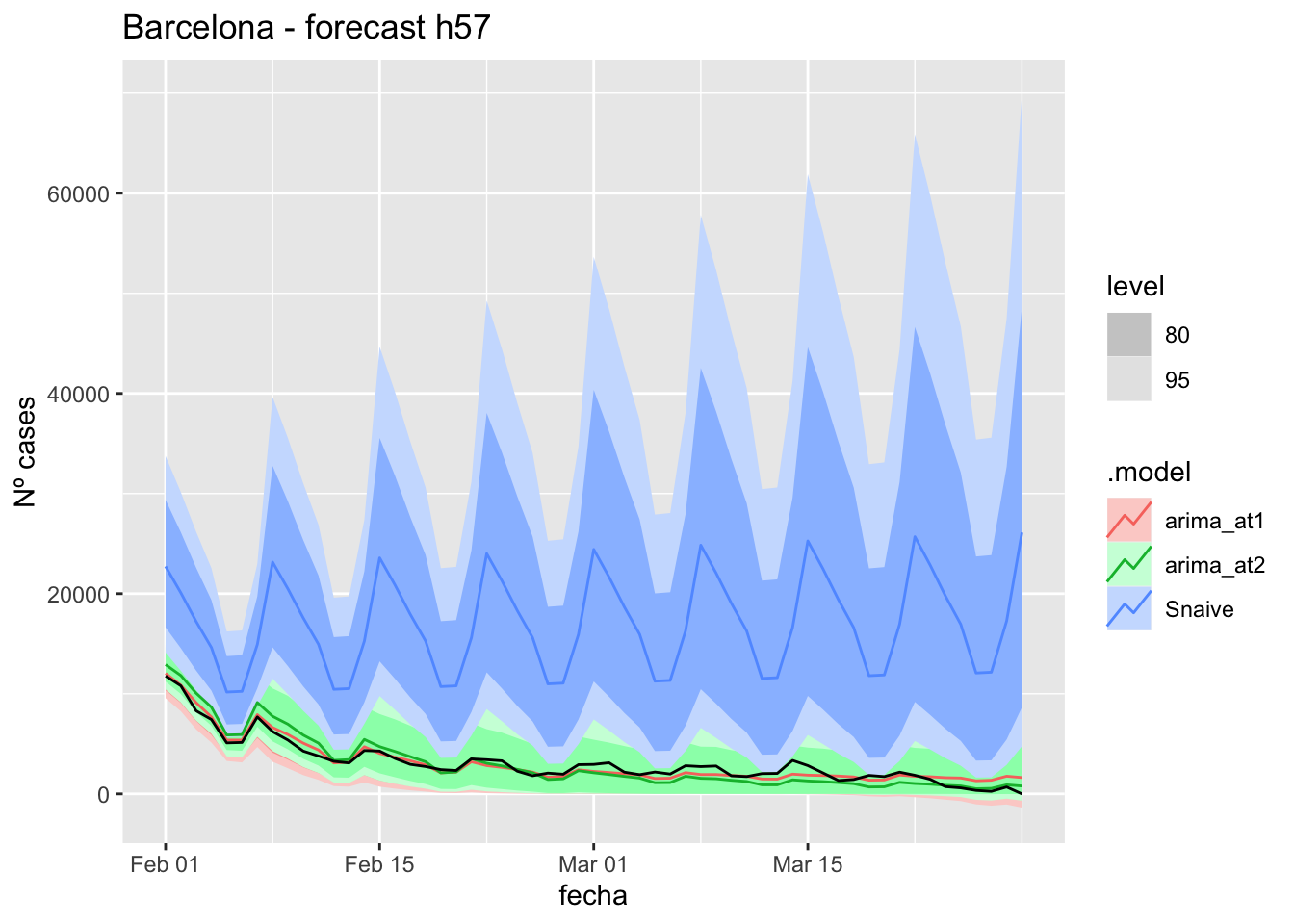
fc_h57 %>%
autoplot(
data_Barcelona %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(57) &
fecha > as.Date("2022-01-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Barcelona - forecast h57")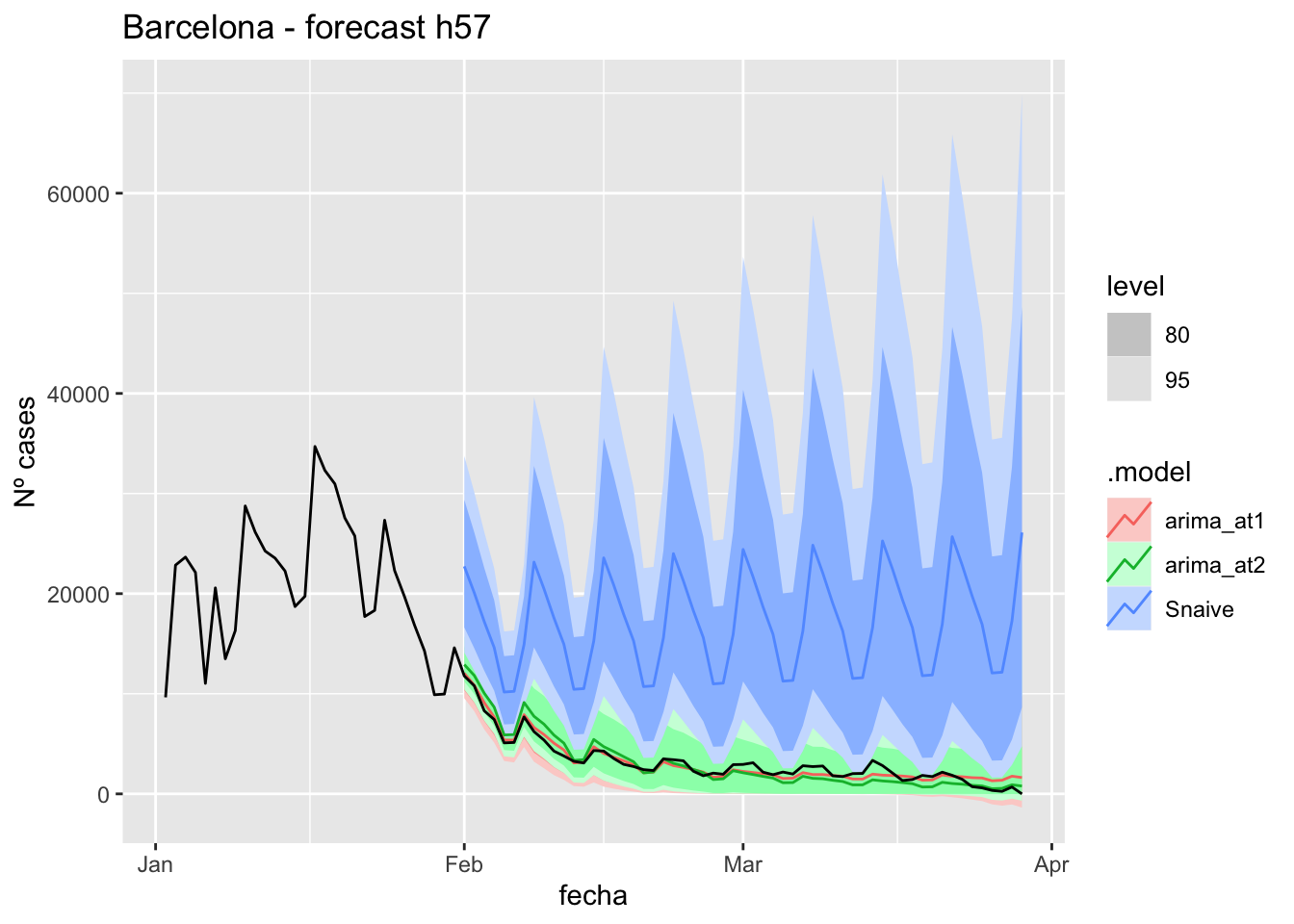
# Accuracy
fabletools::accuracy(fc_h57, data_Barcelona)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_at1 Barcelona Test -2.65e0 598. 482. -Inf Inf 0.605 0.305 0.689
2 arima_at2 Barcelona Test 6.08e1 912. 777. -Inf Inf 0.976 0.466 0.848
3 Snaive Barcelona Test -1.39e4 14803. 13931. -Inf Inf 17.5 7.56 0.580Madrid
data_Madrid <- covid_data %>%
filter(provincia == "Madrid") %>%
filter(fecha > as.Date("2020-06-14", format = "%Y-%m-%d")) %>%
select(provincia, fecha, num_casos, tmed, mob_grocery_pharmacy, mob_parks, mob_residential, mob_retail_recreation, mob_transit_stations, mob_workplaces) %>%
drop_na() %>%
as_tsibble(key = provincia, index = fecha)
data_Madrid# A tsibble: 653 x 10 [1D]
# Key: provincia [1]
provincia fecha num_casos tmed mob_grocery_pharmacy mob_parks
<chr> <date> <dbl> <dbl> <dbl> <dbl>
1 Madrid 2020-06-15 153 20.7 -8 34
2 Madrid 2020-06-16 91 21 -8 21
3 Madrid 2020-06-17 93 20.2 -5 34
4 Madrid 2020-06-18 85 21.9 -7 30
5 Madrid 2020-06-19 78 22.6 -9 22
6 Madrid 2020-06-20 67 23.4 -12 25
7 Madrid 2020-06-21 42 25 -13 14
8 Madrid 2020-06-22 60 27.3 -8 29
9 Madrid 2020-06-23 68 28.4 -9 23
10 Madrid 2020-06-24 49 28 -7 22
# … with 643 more rows, and 4 more variables: mob_residential <dbl>,
# mob_retail_recreation <dbl>, mob_transit_stations <dbl>,
# mob_workplaces <dbl>data_Madrid_train <- data_Madrid %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d"))
summary(data_Madrid_train$fecha) Min. 1st Qu. Median Mean 3rd Qu. Max.
"2020-06-15" "2020-11-10" "2021-04-08" "2021-04-08" "2021-09-04" "2022-01-31" data_Madrid_test <- data_Madrid %>%
filter(fecha > as.Date("2022-01-31", format = "%Y-%m-%d"))
summary(data_Madrid_test$fecha) Min. 1st Qu. Median Mean 3rd Qu. Max.
"2022-02-01" "2022-02-15" "2022-03-01" "2022-03-01" "2022-03-15" "2022-03-29" # Lamba for num_casos
lambda_Madrid_casos <- data_asturias %>%
features(num_casos, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Madrid_casos[1] 0.2278229# Lamba for average temp
lambda_Madrid_tmed <- data_asturias %>%
features(tmed, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Madrid_tmed[1] 1.056074# Lamba for mobility
lambda_Madrid_grocery <- data_asturias %>%
features(mob_grocery_pharmacy, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Madrid_grocery[1] 1.377952lambda_Madrid_parks <- data_asturias %>%
features(mob_parks, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Madrid_parks[1] 0.7188469lambda_Madrid_resd <- data_asturias %>%
features(mob_residential, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Madrid_resd[1] 1.000064lambda_Madrid_retail <- data_asturias %>%
features(mob_retail_recreation, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Madrid_retail[1] 1.058286lambda_Madrid_transit <- data_asturias %>%
features(mob_transit_stations, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Madrid_transit[1] 1.116829lambda_Madrid_work <- data_asturias %>%
features(mob_workplaces, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Madrid_work[1] 1.999927fit_model <- data_Madrid_train %>%
model(
arima_at1 = ARIMA(box_cox(num_casos, lambda_Madrid_casos) ~
box_cox(tmed, lambda_Madrid_tmed) +
box_cox(mob_grocery_pharmacy, lambda_Madrid_grocery) +
box_cox(mob_parks, lambda_Madrid_parks) +
box_cox(mob_residential, lambda_Madrid_resd) +
box_cox(mob_retail_recreation, lambda_Madrid_retail) +
box_cox(mob_transit_stations, lambda_Madrid_transit) +
box_cox(mob_workplaces, lambda_Madrid_work)
),
arima_at2 = ARIMA(box_cox(num_casos, lambda_Madrid_casos) ~
box_cox(tmed, lambda_Madrid_tmed) +
box_cox(mob_grocery_pharmacy, lambda_Madrid_grocery) +
box_cox(mob_parks, lambda_Madrid_parks) +
box_cox(mob_residential, lambda_Madrid_resd) +
box_cox(mob_retail_recreation, lambda_Madrid_retail) +
box_cox(mob_transit_stations, lambda_Madrid_transit) +
box_cox(mob_workplaces, lambda_Madrid_work),
stepwise = FALSE, approx = FALSE),
Snaive = SNAIVE(box_cox(num_casos, lambda_Madrid_casos))
)
# Show and report model
fit_model# A mable: 1 x 4
# Key: provincia [1]
provincia arima_at1
<chr> <model>
1 Madrid <LM w/ ARIMA(1,1,1)(1,0,2)[7] errors>
# … with 2 more variables: arima_at2 <model>, Snaive <model>glance(fit_model)# A tibble: 3 × 9
provincia .model sigma2 log_lik AIC AICc BIC ar_roots ma_roots
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <list> <list>
1 Madrid arima_at1 1.18 -892. 1809. 1810. 1866. <cpl [8]> <cpl [15]>
2 Madrid arima_at2 1.17 -889. 1804. 1804. 1861. <cpl [8]> <cpl [9]>
3 Madrid Snaive 5.10 NA NA NA NA <NULL> <NULL> fit_model %>%
pivot_longer(-provincia,
names_to = ".model",
values_to = "orders") %>%
left_join(glance(fit_model), by = c("provincia", ".model")) %>%
arrange(AICc) %>%
select(.model:BIC)# A mable: 3 x 7
# Key: .model [3]
.model orders sigma2 log_lik AIC AICc BIC
<chr> <model> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_… <LM w/ ARIMA(1,1,2)(1,0,1)[7] errors> 1.17 -889. 1804. 1804. 1861.
2 arima_… <LM w/ ARIMA(1,1,1)(1,0,2)[7] errors> 1.18 -892. 1809. 1810. 1866.
3 Snaive <SNAIVE> 5.10 NA NA NA NA # We use a Ljung-Box test >> large p-value, confirms residuals are similar / considered to white noise
fit_model %>%
select(Snaive) %>%
gg_tsresiduals()
fit_model %>%
select(arima_at1) %>%
gg_tsresiduals()
fit_model %>%
select(arima_at2) %>%
gg_tsresiduals()
augment(fit_model) %>%
features(.innov, ljung_box, lag=7)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Madrid arima_at1 12.6 0.0823
2 Madrid arima_at2 6.30 0.506
3 Madrid Snaive 1552. 0 augment(fit_model) %>%
features(.innov, ljung_box, lag=14)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Madrid arima_at1 25.5 0.0299
2 Madrid arima_at2 21.3 0.0953
3 Madrid Snaive 2076. 0 augment(fit_model) %>%
features(.innov, ljung_box, lag=21)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Madrid arima_at1 36.2 0.0208
2 Madrid arima_at2 33.0 0.0462
3 Madrid Snaive 2151. 0 augment(fit_model) %>%
features(.innov, ljung_box, lag=57)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Madrid arima_at1 86.2 0.00747
2 Madrid arima_at2 78.0 0.0338
3 Madrid Snaive 2518. 0 # To predict future values of infections, we need to pass the model future/prediction values of the complementary variables tmed and mobility
# We are going to select those from the test dataset
data_fc_h7 <- data_Madrid_test %>%
select(-num_casos) %>%
slice(1:7)
data_fc_h14 <- data_Madrid_test %>%
select(-num_casos) %>%
slice(1:14)
data_fc_h21 <- data_Madrid_test %>%
select(-num_casos) %>%
slice(1:21)
data_fc_h57 <- data_Madrid_test %>%
select(-num_casos) %>%
slice(1:57)# Significant spikes out of 30 is still consistent with white noise
# To be sure, use a Ljung-Box test, which has a large p-value, confirming that
# the residuals are similar to white noise.
# Note that the alternative models also passed this test.
#
# Forecast
fc_h7 <- fabletools::forecast(fit_model, new_data = data_fc_h7)
fc_h14 <- fabletools::forecast(fit_model, new_data = data_fc_h14)
fc_h21 <- fabletools::forecast(fit_model, new_data = data_fc_h21)
fc_h57 <- fabletools::forecast(fit_model, new_data = data_fc_h57)7 days
# Plots
fc_h7 %>%
autoplot(
data_Madrid_test %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(7))
) +
labs(y = "Nº cases", title = "Madrid - forecast h7")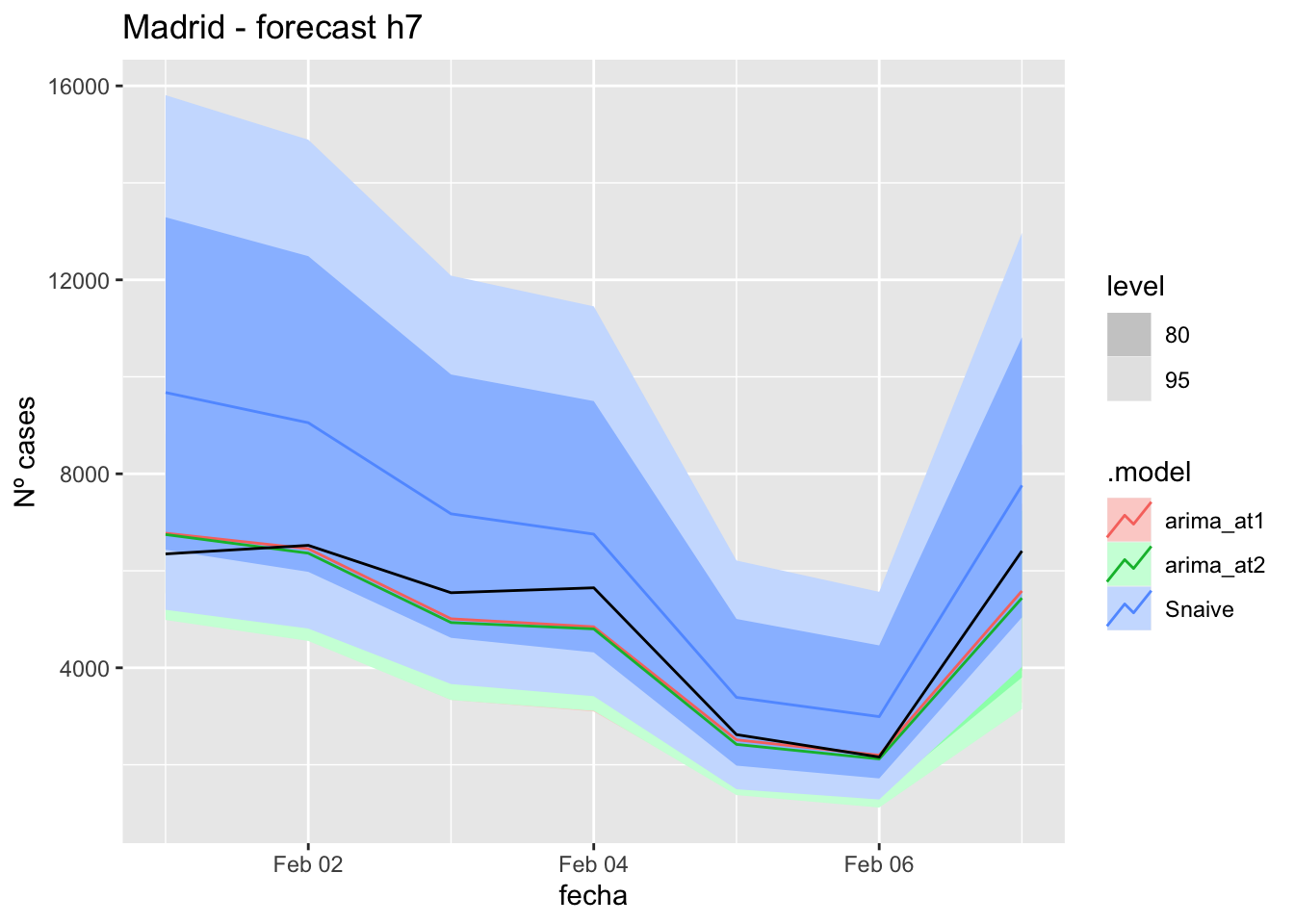
fc_h7 %>%
autoplot(
data_Madrid %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(7) &
fecha > as.Date("2022-01-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Madrid - forecast h7")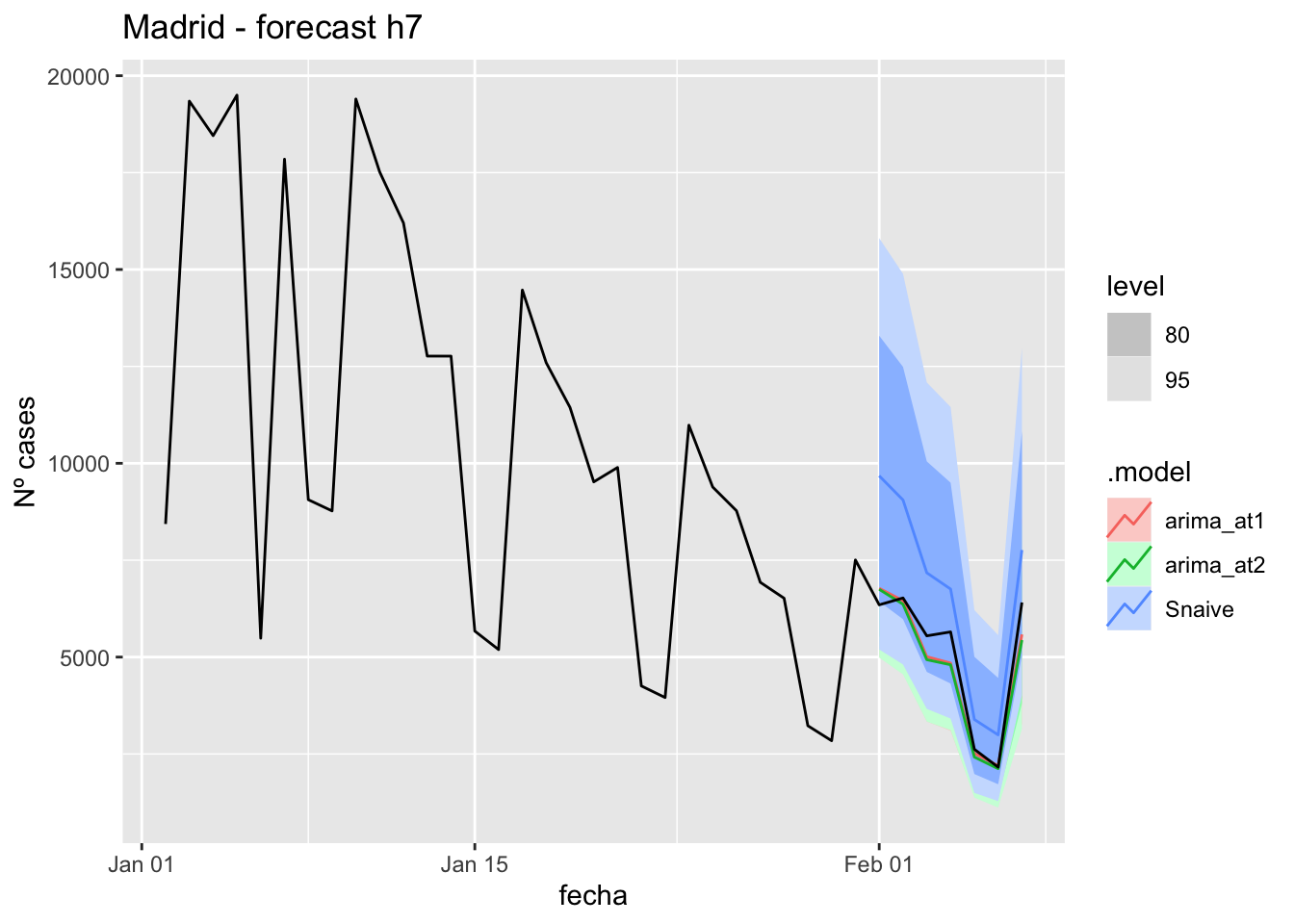
# Accuracy
fabletools::accuracy(fc_h7, data_Madrid)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_at1 Madrid Test 268. 509. 401. 4.78 7.20 0.537 0.285 0.0164
2 arima_at2 Madrid Test 348. 569. 462. 6.71 8.51 0.619 0.319 0.00237
3 Snaive Madrid Test -1649. 1869. 1649. -32.7 32.7 2.21 1.05 0.537 14 days
# Plots
fc_h14 %>%
autoplot(
data_Madrid_test %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(14))
) +
labs(y = "Nº cases", title = "Madrid - forecast h14")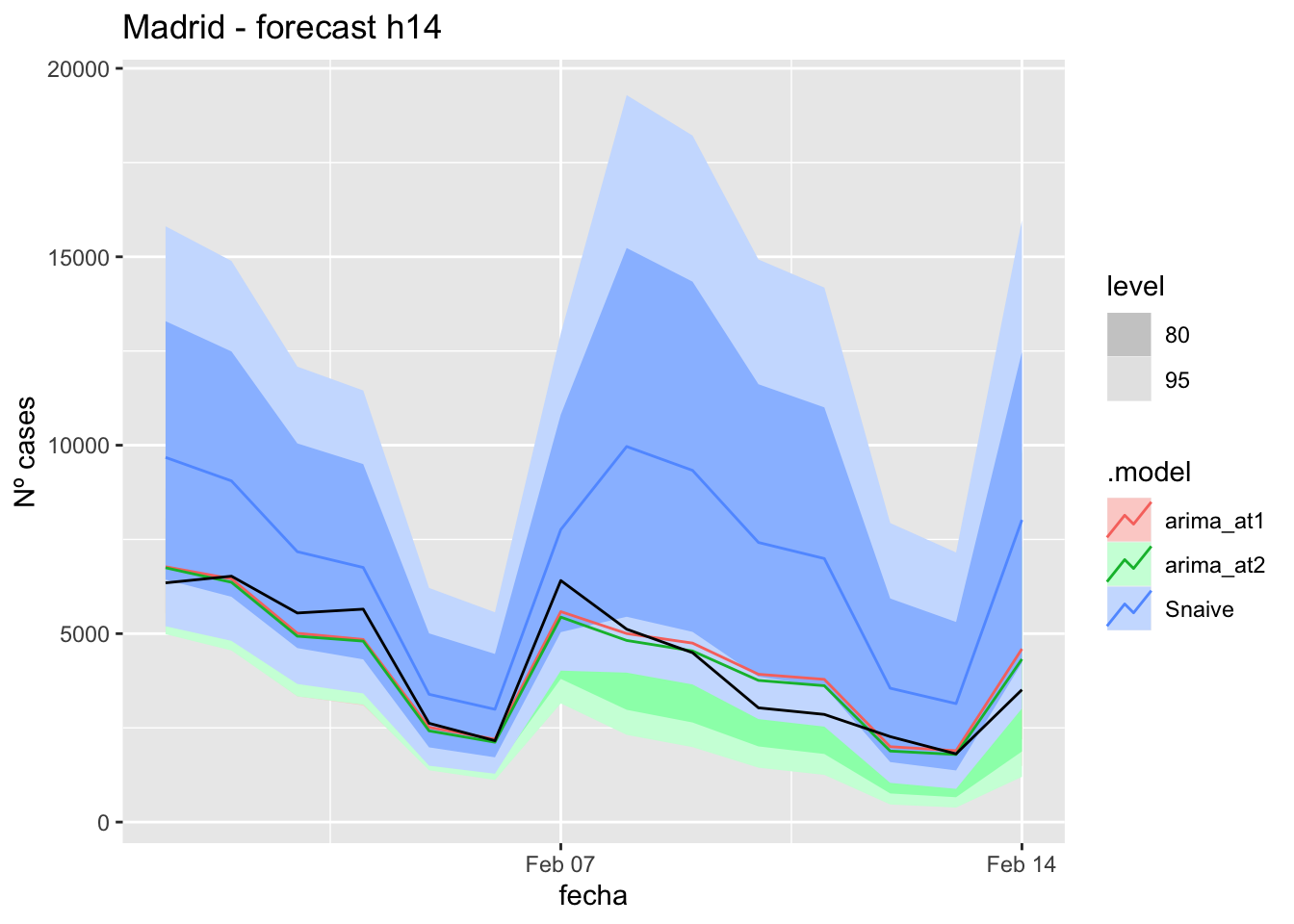
fc_h14 %>%
autoplot(
data_Madrid %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(14) &
fecha > as.Date("2022-01-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Madrid - forecast h14")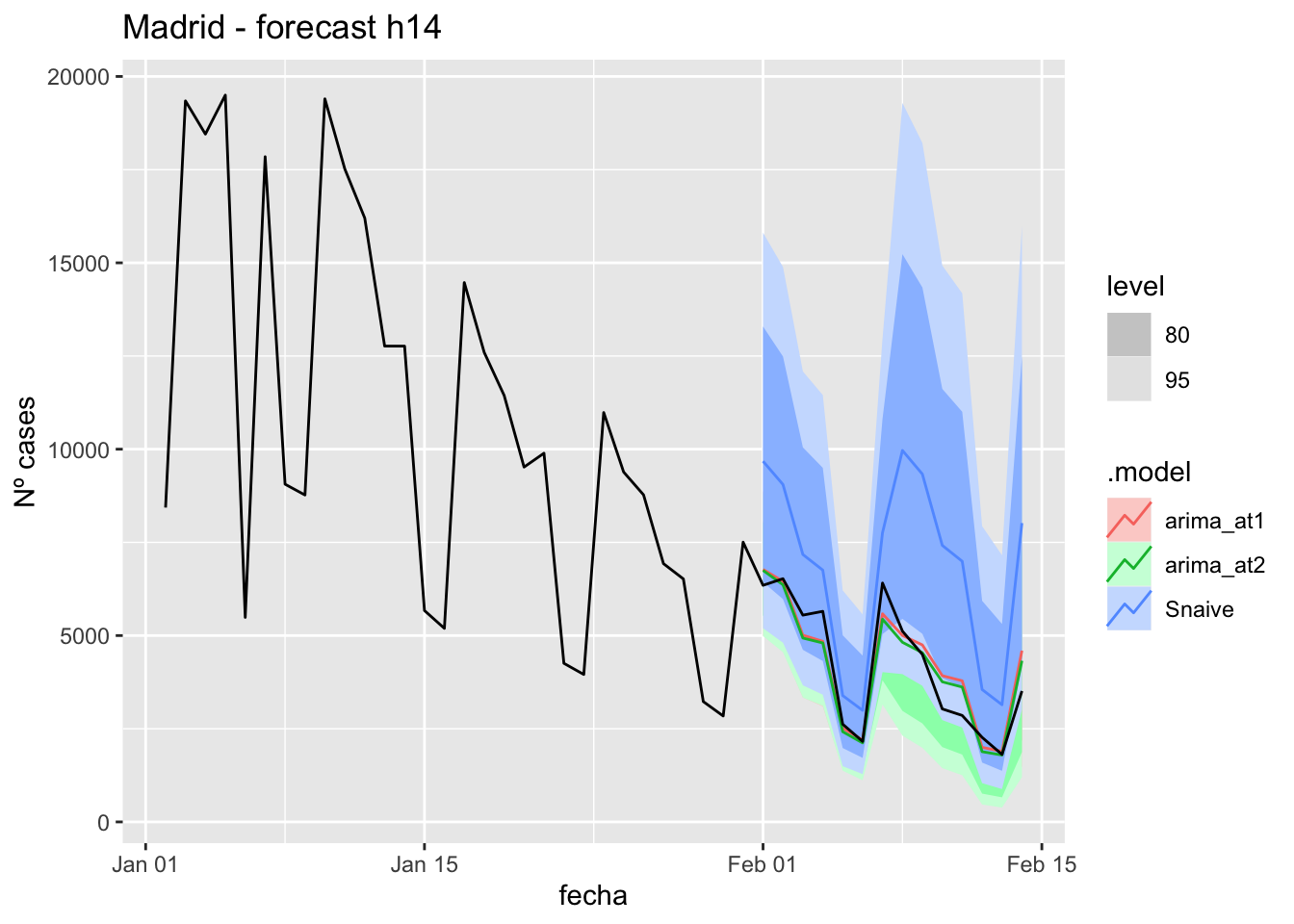
# Accuracy
fabletools::accuracy(fc_h14, data_Madrid)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_at1 Madrid Test -70.1 586. 460. -3.97 12.0 0.615 0.328 0.309
2 arima_at2 Madrid Test 56.2 553. 449. -0.301 11.3 0.601 0.309 0.289
3 Snaive Madrid Test -2634. 3064. 2634. -70.0 70.0 3.53 1.72 0.46421 days
# Plots
fc_h21 %>%
autoplot(
data_Madrid_test %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(21))
) +
labs(y = "Nº cases", title = "Madrid - forecast h21")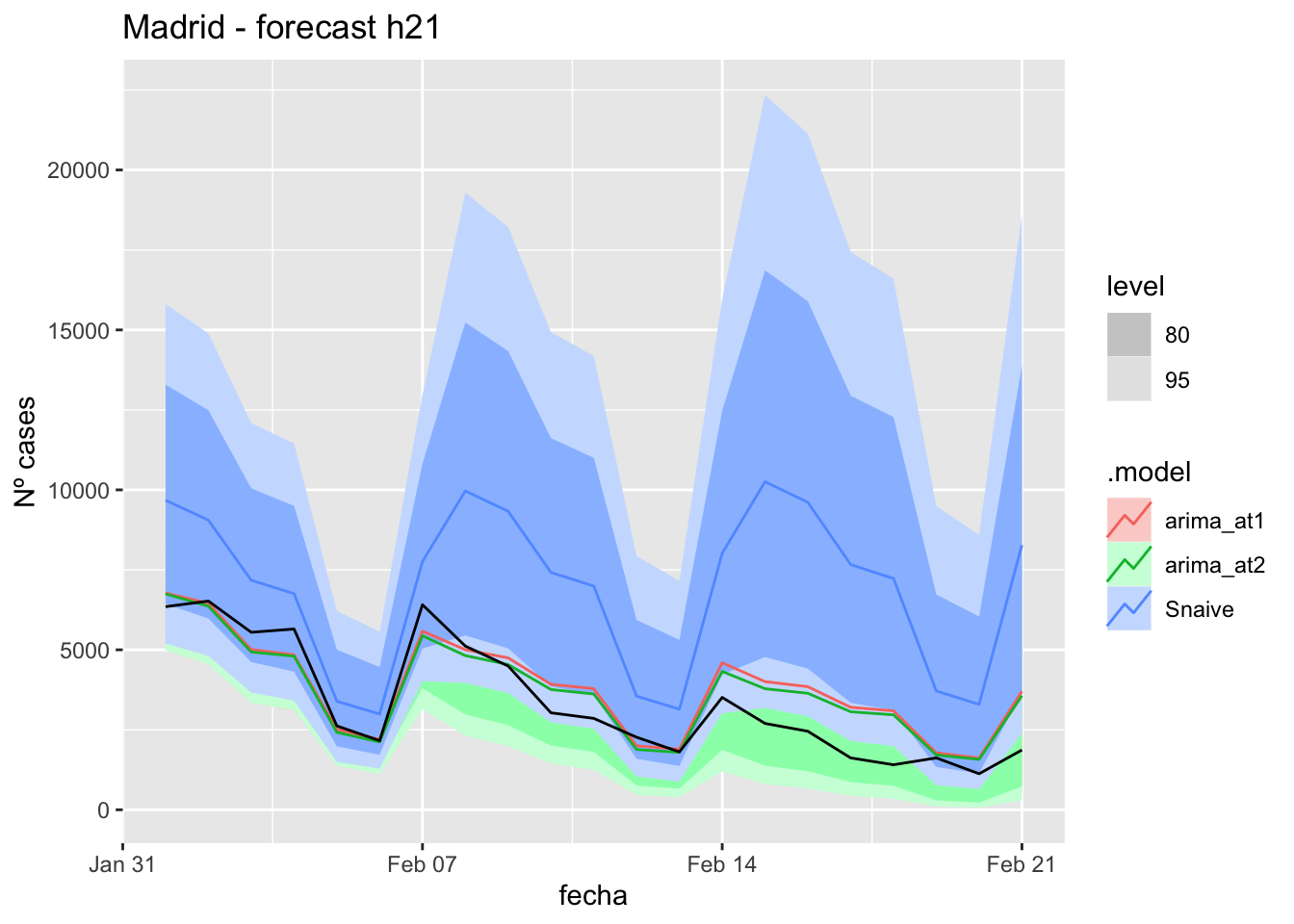
fc_h21 %>%
autoplot(
data_Madrid %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(21) &
fecha > as.Date("2022-01-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Madrid - forecast h21")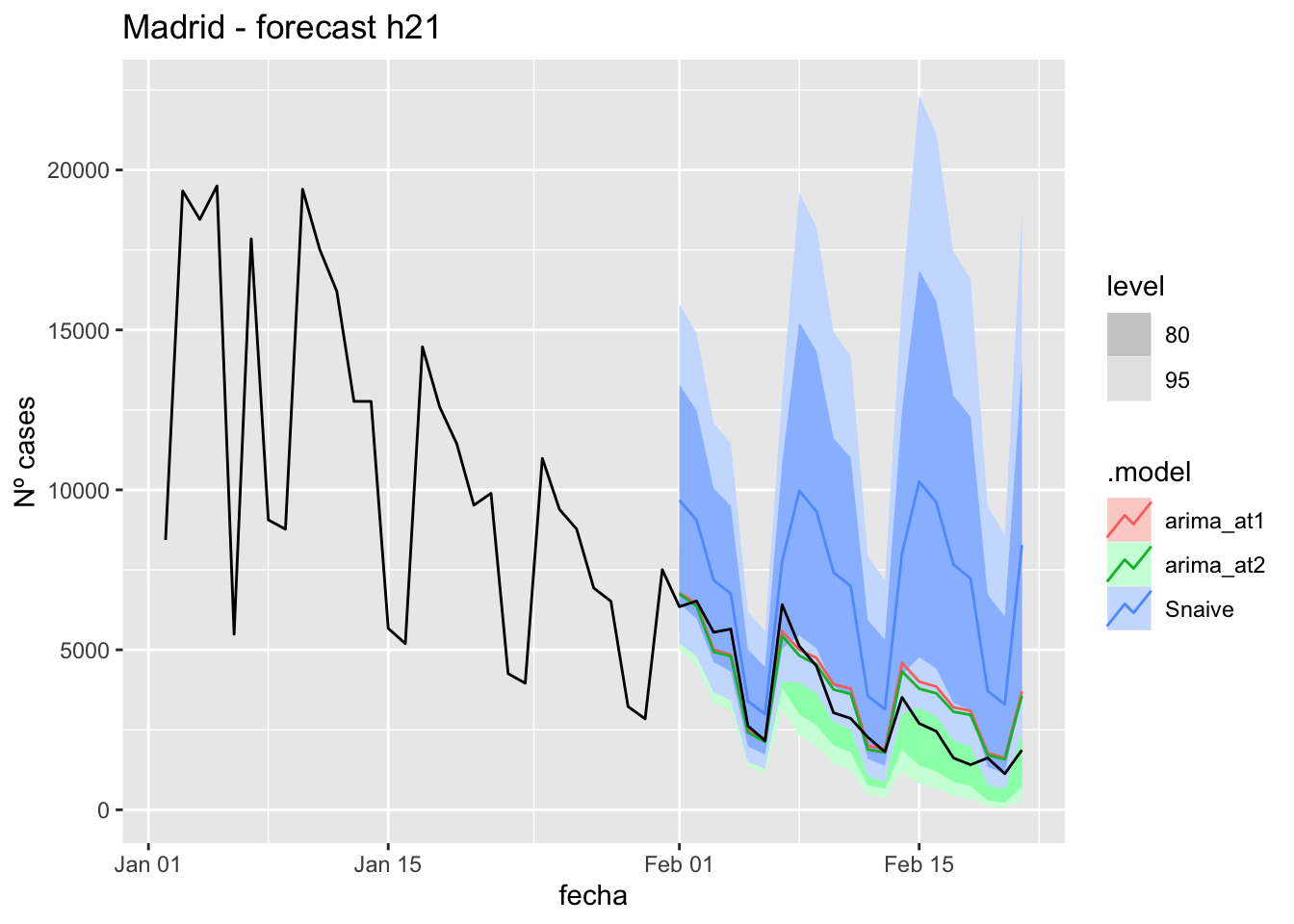
# Accuracy
fabletools::accuracy(fc_h21, data_Madrid)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_at1 Madrid Test -449. 911. 709. -25.2 30.6 0.949 0.510 0.559
2 arima_at2 Madrid Test -320. 830. 657. -20.4 27.8 0.880 0.465 0.555
3 Snaive Madrid Test -3530. 4142. 3530. -143. 143. 4.73 2.32 0.57657 days
# Plots
fc_h57 %>%
autoplot(
data_Madrid_test %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(57))
) +
labs(y = "Nº cases", title = "Madrid - forecast h57")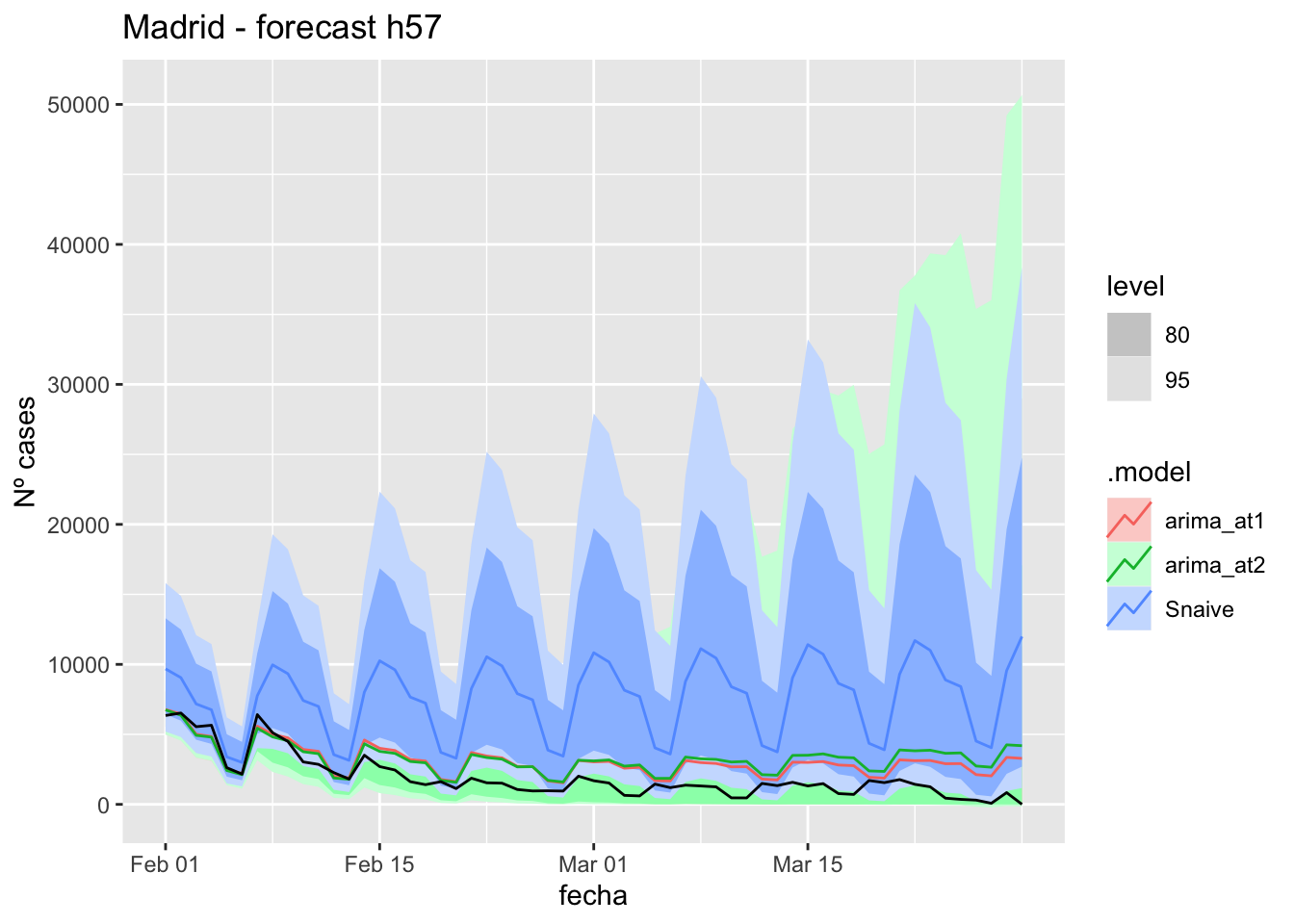
fc_h57 %>%
autoplot(
data_Madrid %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(57) &
fecha > as.Date("2022-01-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Madrid - forecast h57")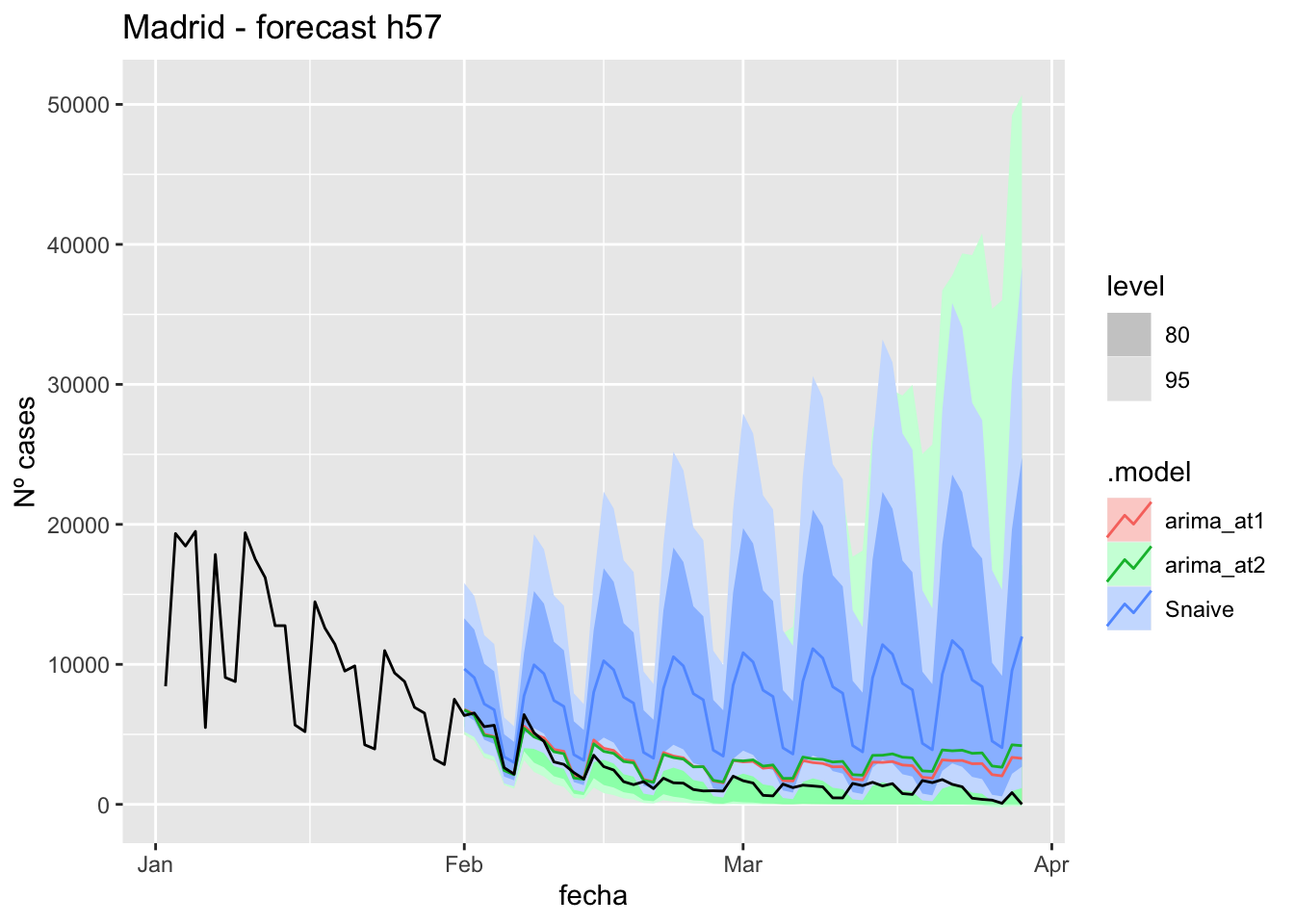
# Accuracy
fabletools::accuracy(fc_h57, data_Madrid)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_at1 Madrid Test -1153. 1478. 1249. -1126. 1128. 1.67 0.828 0.660
2 arima_at2 Madrid Test -1337. 1762. 1461. -1430. 1433. 1.96 0.986 0.741
3 Snaive Madrid Test -5635. 6384. 5635. -4097. 4097. 7.54 3.57 0.622Malaga
data_Malaga <- covid_data %>%
filter(provincia == "Málaga") %>%
filter(fecha > as.Date("2020-06-14", format = "%Y-%m-%d")) %>%
select(provincia, fecha, num_casos, tmed, mob_grocery_pharmacy, mob_parks, mob_residential, mob_retail_recreation, mob_transit_stations, mob_workplaces) %>%
drop_na() %>%
as_tsibble(key = provincia, index = fecha)
data_Malaga# A tsibble: 653 x 10 [1D]
# Key: provincia [1]
provincia fecha num_casos tmed mob_grocery_pharmacy mob_parks
<chr> <date> <dbl> <dbl> <dbl> <dbl>
1 Málaga 2020-06-15 1 27.2 -16 -9
2 Málaga 2020-06-16 1 27.8 -13 -4
3 Málaga 2020-06-17 2 26.9 -13 1
4 Málaga 2020-06-18 2 21.6 -12 3
5 Málaga 2020-06-19 1 24.7 -12 2
6 Málaga 2020-06-20 4 22.6 -14 8
7 Málaga 2020-06-21 4 22.7 -16 -3
8 Málaga 2020-06-22 6 25 -13 1
9 Málaga 2020-06-23 8 25.6 -10 18
10 Málaga 2020-06-24 65 24.5 -13 12
# … with 643 more rows, and 4 more variables: mob_residential <dbl>,
# mob_retail_recreation <dbl>, mob_transit_stations <dbl>,
# mob_workplaces <dbl>data_Malaga_train <- data_Malaga %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d"))
summary(data_Malaga_train$fecha) Min. 1st Qu. Median Mean 3rd Qu. Max.
"2020-06-15" "2020-11-10" "2021-04-08" "2021-04-08" "2021-09-04" "2022-01-31" data_Malaga_test <- data_Malaga %>%
filter(fecha > as.Date("2022-01-31", format = "%Y-%m-%d"))
summary(data_Malaga_test$fecha) Min. 1st Qu. Median Mean 3rd Qu. Max.
"2022-02-01" "2022-02-15" "2022-03-01" "2022-03-01" "2022-03-15" "2022-03-29" # Lamba for num_casos
lambda_Malaga_casos <- data_asturias %>%
features(num_casos, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Malaga_casos[1] 0.2278229# Lamba for average temp
lambda_Malaga_tmed <- data_asturias %>%
features(tmed, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Malaga_tmed[1] 1.056074# Lamba for mobility
lambda_Malaga_grocery <- data_asturias %>%
features(mob_grocery_pharmacy, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Malaga_grocery[1] 1.377952lambda_Malaga_parks <- data_asturias %>%
features(mob_parks, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Malaga_parks[1] 0.7188469lambda_Malaga_resd <- data_asturias %>%
features(mob_residential, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Malaga_resd[1] 1.000064lambda_Malaga_retail <- data_asturias %>%
features(mob_retail_recreation, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Malaga_retail[1] 1.058286lambda_Malaga_transit <- data_asturias %>%
features(mob_transit_stations, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Malaga_transit[1] 1.116829lambda_Malaga_work <- data_asturias %>%
features(mob_workplaces, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Malaga_work[1] 1.999927fit_model <- data_Malaga_train %>%
model(
arima_at1 = ARIMA(box_cox(num_casos, lambda_Malaga_casos) ~
box_cox(tmed, lambda_Malaga_tmed) +
box_cox(mob_grocery_pharmacy, lambda_Malaga_grocery) +
box_cox(mob_parks, lambda_Malaga_parks) +
box_cox(mob_residential, lambda_Malaga_resd) +
box_cox(mob_retail_recreation, lambda_Malaga_retail) +
box_cox(mob_transit_stations, lambda_Malaga_transit) +
box_cox(mob_workplaces, lambda_Malaga_work)
),
arima_at2 = ARIMA(box_cox(num_casos, lambda_Malaga_casos) ~
box_cox(tmed, lambda_Malaga_tmed) +
box_cox(mob_grocery_pharmacy, lambda_Malaga_grocery) +
box_cox(mob_parks, lambda_Malaga_parks) +
box_cox(mob_residential, lambda_Malaga_resd) +
box_cox(mob_retail_recreation, lambda_Malaga_retail) +
box_cox(mob_transit_stations, lambda_Malaga_transit) +
box_cox(mob_workplaces, lambda_Malaga_work),
stepwise = FALSE, approx = FALSE),
Snaive = SNAIVE(box_cox(num_casos, lambda_Malaga_casos))
)
# Show and report model
fit_model# A mable: 1 x 4
# Key: provincia [1]
provincia arima_at1
<chr> <model>
1 Málaga <LM w/ ARIMA(4,1,0)(2,0,0)[7] errors>
# … with 2 more variables: arima_at2 <model>, Snaive <model>glance(fit_model)# A tibble: 3 × 9
provincia .model sigma2 log_lik AIC AICc BIC ar_roots ma_roots
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <list> <list>
1 Málaga arima_at1 0.568 -671. 1369. 1370. 1431. <cpl [18]> <cpl [0]>
2 Málaga arima_at2 0.550 -662. 1352. 1353. 1414. <cpl [7]> <cpl [11]>
3 Málaga Snaive 2.32 NA NA NA NA <NULL> <NULL> fit_model %>%
pivot_longer(-provincia,
names_to = ".model",
values_to = "orders") %>%
left_join(glance(fit_model), by = c("provincia", ".model")) %>%
arrange(AICc) %>%
select(.model:BIC)# A mable: 3 x 7
# Key: .model [3]
.model orders sigma2 log_lik AIC AICc BIC
<chr> <model> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_… <LM w/ ARIMA(0,1,4)(1,0,1)[7] errors> 0.550 -662. 1352. 1353. 1414.
2 arima_… <LM w/ ARIMA(4,1,0)(2,0,0)[7] errors> 0.568 -671. 1369. 1370. 1431.
3 Snaive <SNAIVE> 2.32 NA NA NA NA # We use a Ljung-Box test >> large p-value, confirms residuals are similar / considered to white noise
fit_model %>%
select(Snaive) %>%
gg_tsresiduals()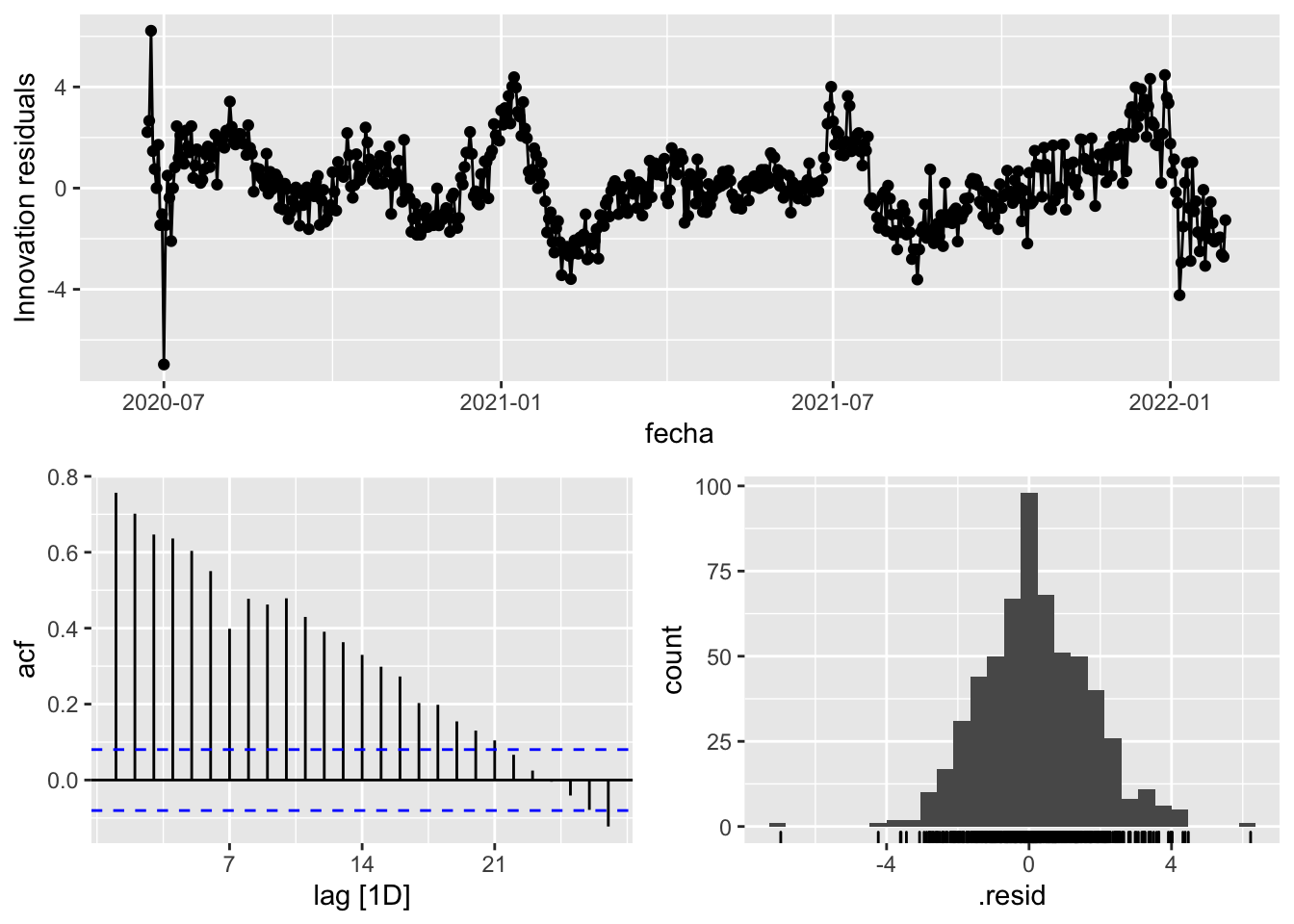
fit_model %>%
select(arima_at1) %>%
gg_tsresiduals()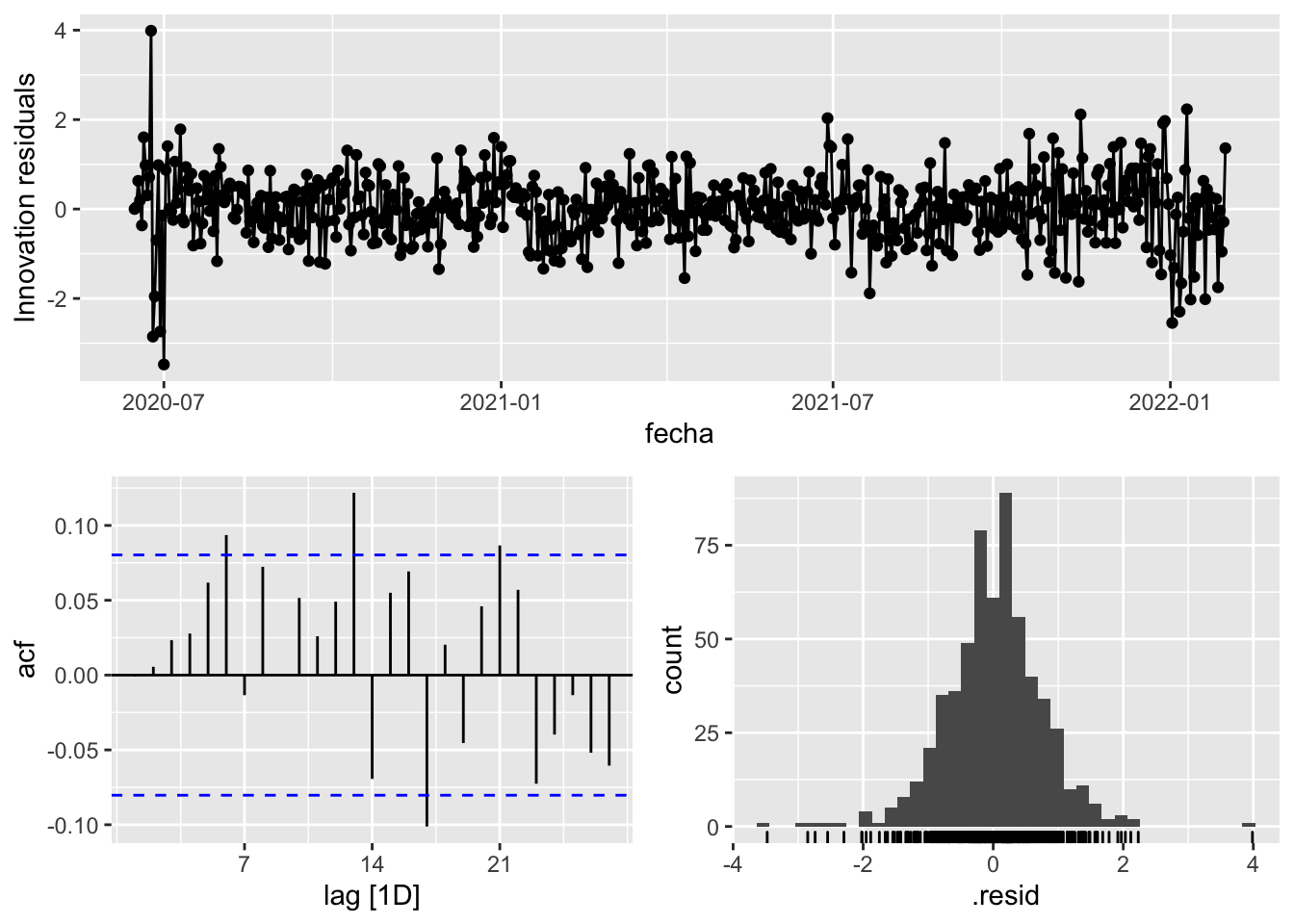
fit_model %>%
select(arima_at2) %>%
gg_tsresiduals()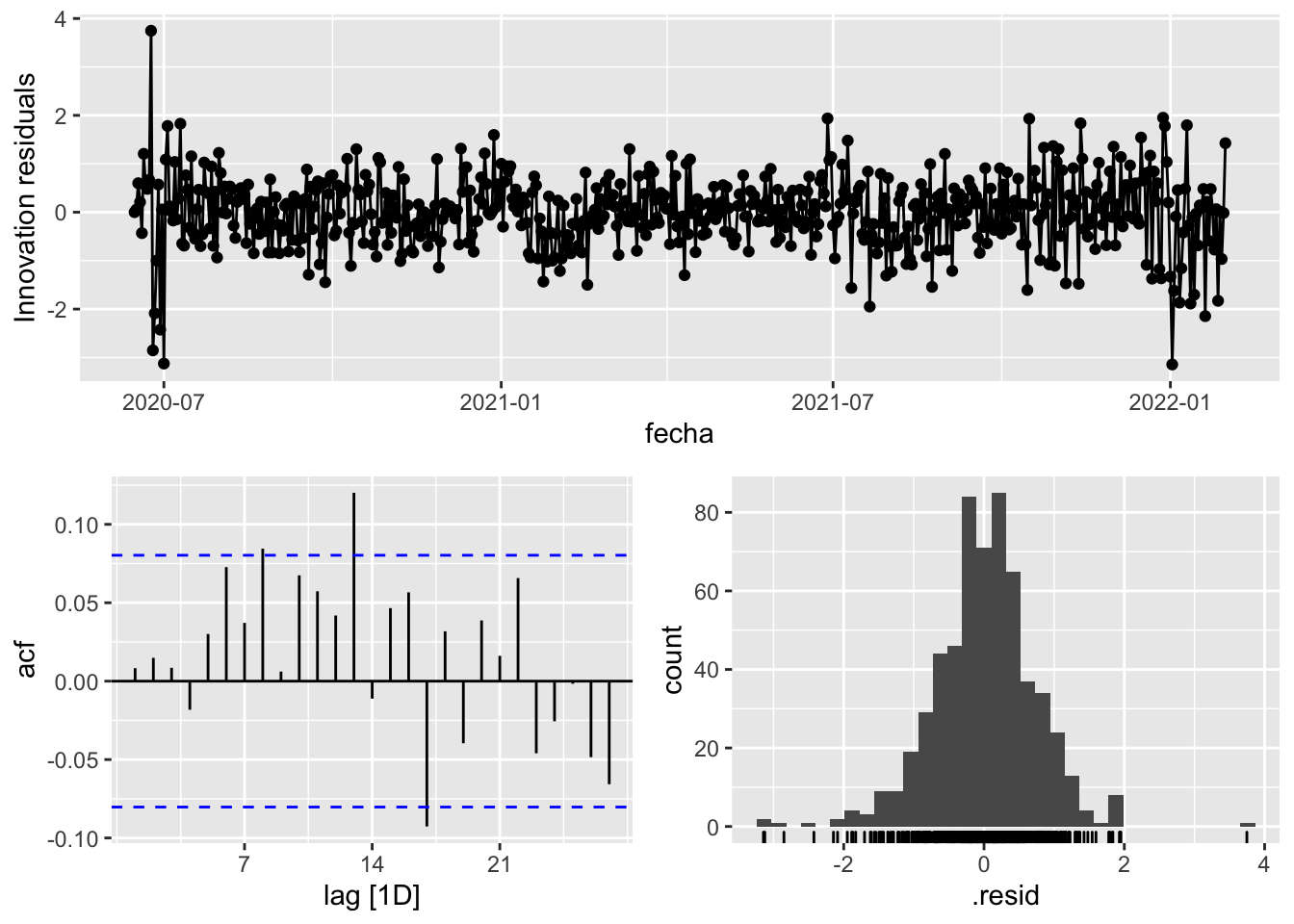
augment(fit_model) %>%
features(.innov, ljung_box, lag=7)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Málaga arima_at1 8.51 0.290
2 Málaga arima_at2 4.99 0.661
3 Málaga Snaive 1613. 0 augment(fit_model) %>%
features(.innov, ljung_box, lag=14)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Málaga arima_at1 27.2 0.0181
2 Málaga arima_at2 24.1 0.0447
3 Málaga Snaive 2365. 0 augment(fit_model) %>%
features(.innov, ljung_box, lag=21)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Málaga arima_at1 45.8 0.00136
2 Málaga arima_at2 35.4 0.0258
3 Málaga Snaive 2545. 0 augment(fit_model) %>%
features(.innov, ljung_box, lag=57)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Málaga arima_at1 99.1 0.000463
2 Málaga arima_at2 87.6 0.00568
3 Málaga Snaive 3404. 0 # To predict future values of infections, we need to pass the model future/prediction values of the complementary variables tmed and mobility
# We are going to select those from the test dataset
data_fc_h7 <- data_Malaga_test %>%
select(-num_casos) %>%
slice(1:7)
data_fc_h14 <- data_Malaga_test %>%
select(-num_casos) %>%
slice(1:14)
data_fc_h21 <- data_Malaga_test %>%
select(-num_casos) %>%
slice(1:21)
data_fc_h57 <- data_Malaga_test %>%
select(-num_casos) %>%
slice(1:57)# Significant spikes out of 30 is still consistent with white noise
# To be sure, use a Ljung-Box test, which has a large p-value, confirming that
# the residuals are similar to white noise.
# Note that the alternative models also passed this test.
#
# Forecast
fc_h7 <- fabletools::forecast(fit_model, new_data = data_fc_h7)
fc_h14 <- fabletools::forecast(fit_model, new_data = data_fc_h14)
fc_h21 <- fabletools::forecast(fit_model, new_data = data_fc_h21)
fc_h57 <- fabletools::forecast(fit_model, new_data = data_fc_h57)7 days
# Plots
fc_h7 %>%
autoplot(
data_Malaga_test %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(7))
) +
labs(y = "Nº cases", title = "Malaga - forecast h7")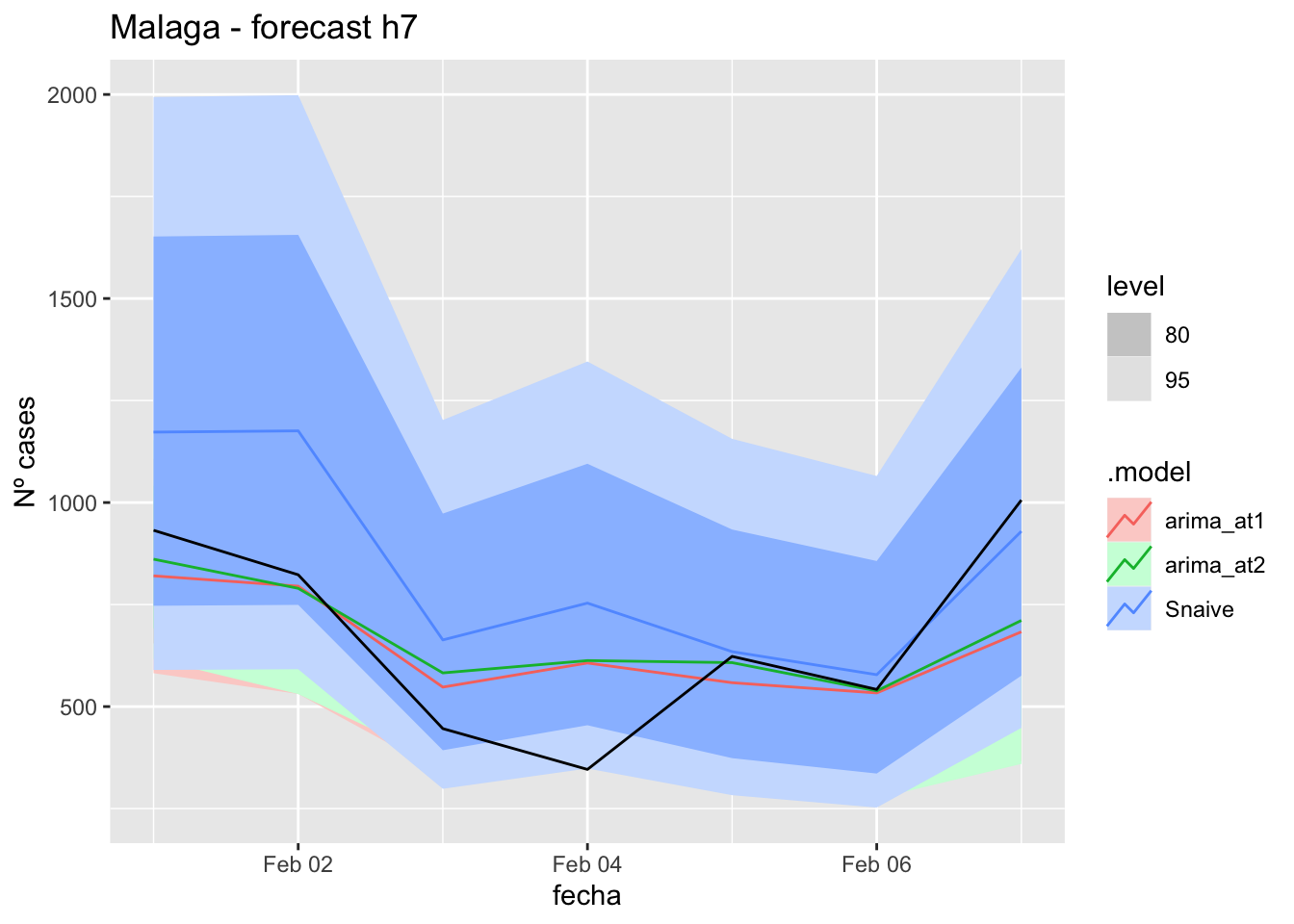
fc_h7 %>%
autoplot(
data_Malaga %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(7) &
fecha > as.Date("2022-01-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Malaga - forecast h7")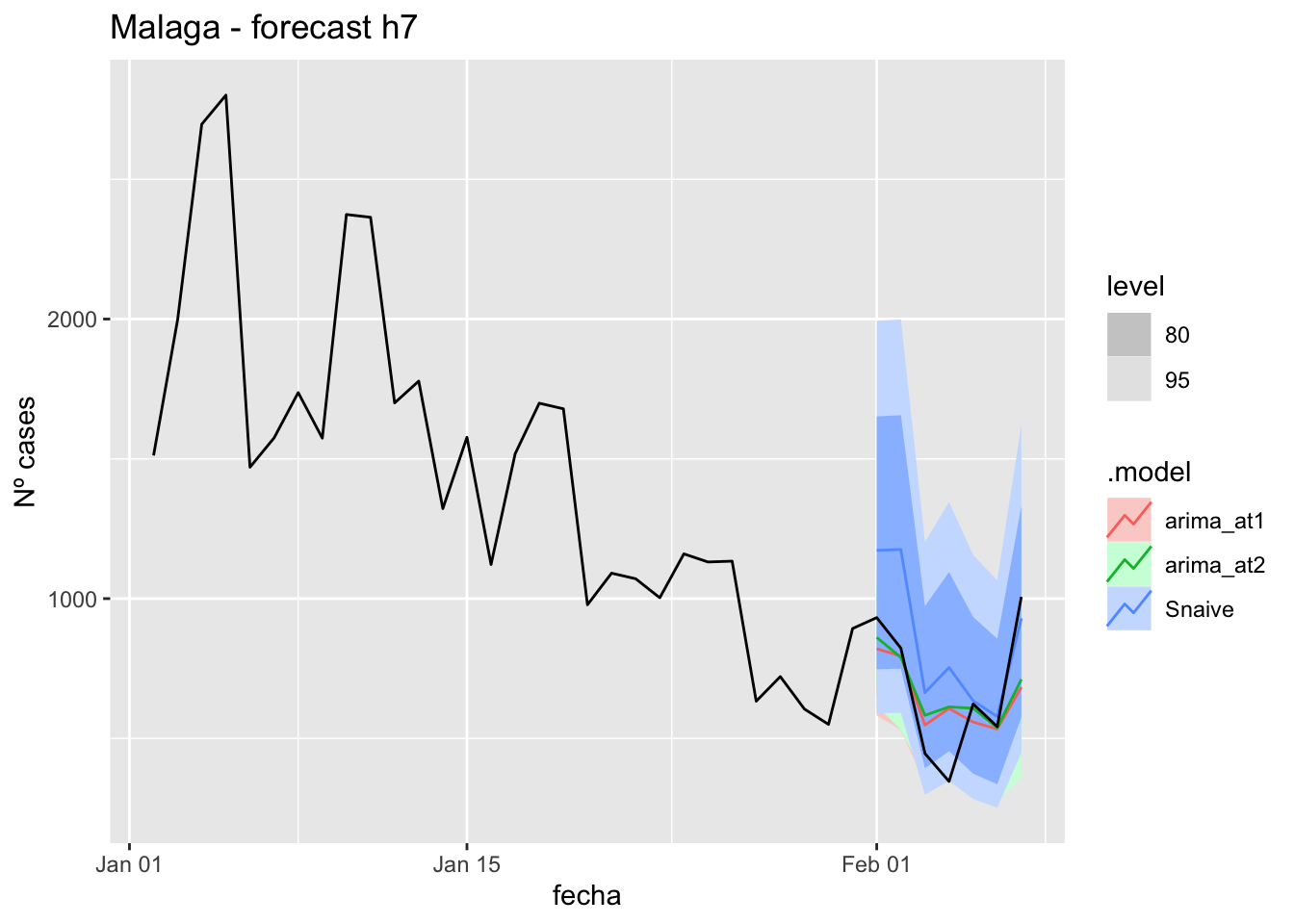
# Accuracy
fabletools::accuracy(fc_h7, data_Malaga)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_at1 Málaga Test 24.7 169. 128. -5.55 22.5 0.971 0.683 0.0978
2 arima_at2 Málaga Test 2.08 162. 117. -9.09 21.7 0.888 0.653 0.175
3 Snaive Málaga Test -170. 240. 192. -33.7 35.9 1.45 0.969 0.246 14 days
# Plots
fc_h14 %>%
autoplot(
data_Malaga_test %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(14))
) +
labs(y = "Nº cases", title = "Malaga - forecast h14")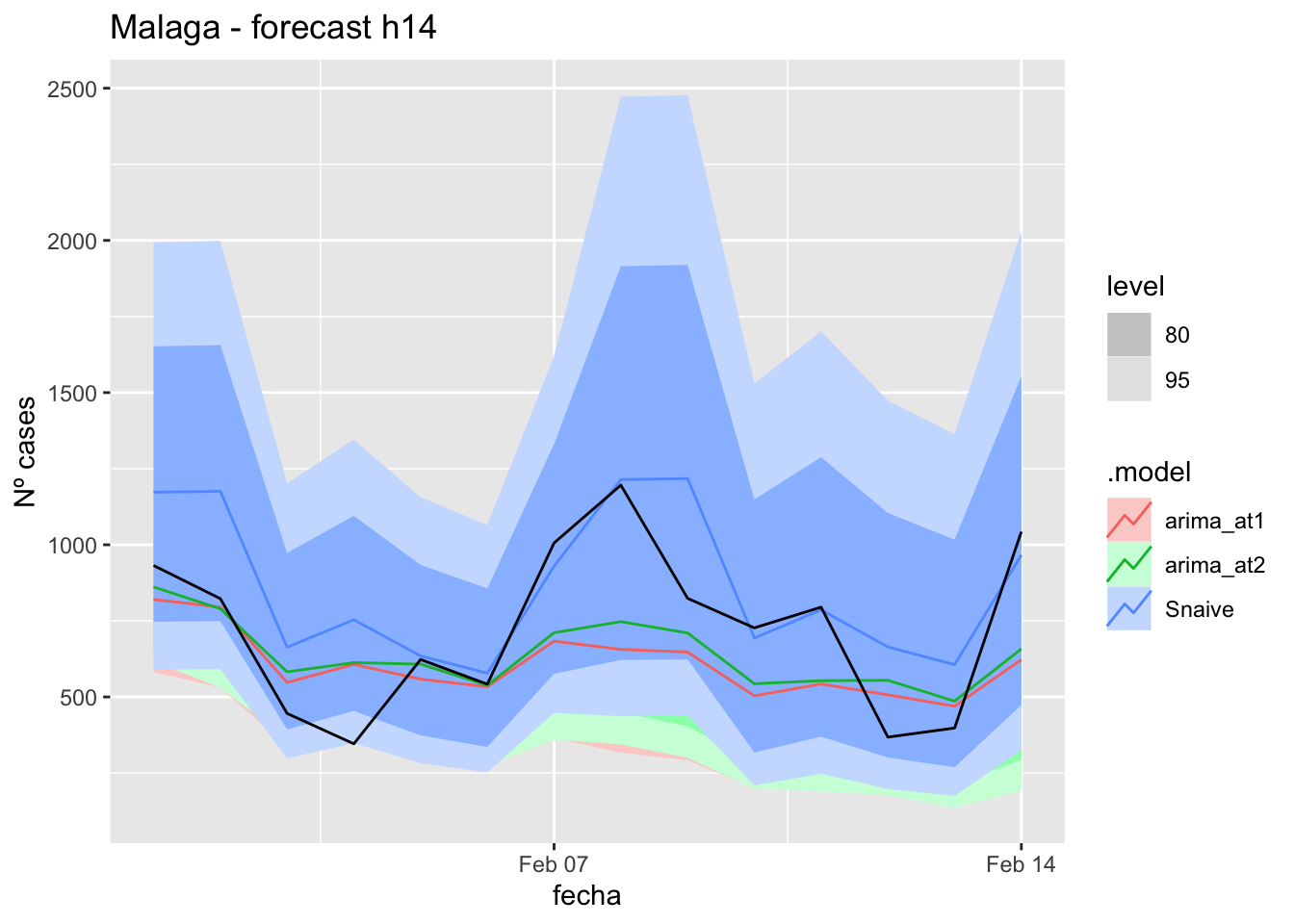
fc_h14 %>%
autoplot(
data_Malaga %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(14) &
fecha > as.Date("2022-01-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Malaga - forecast h14")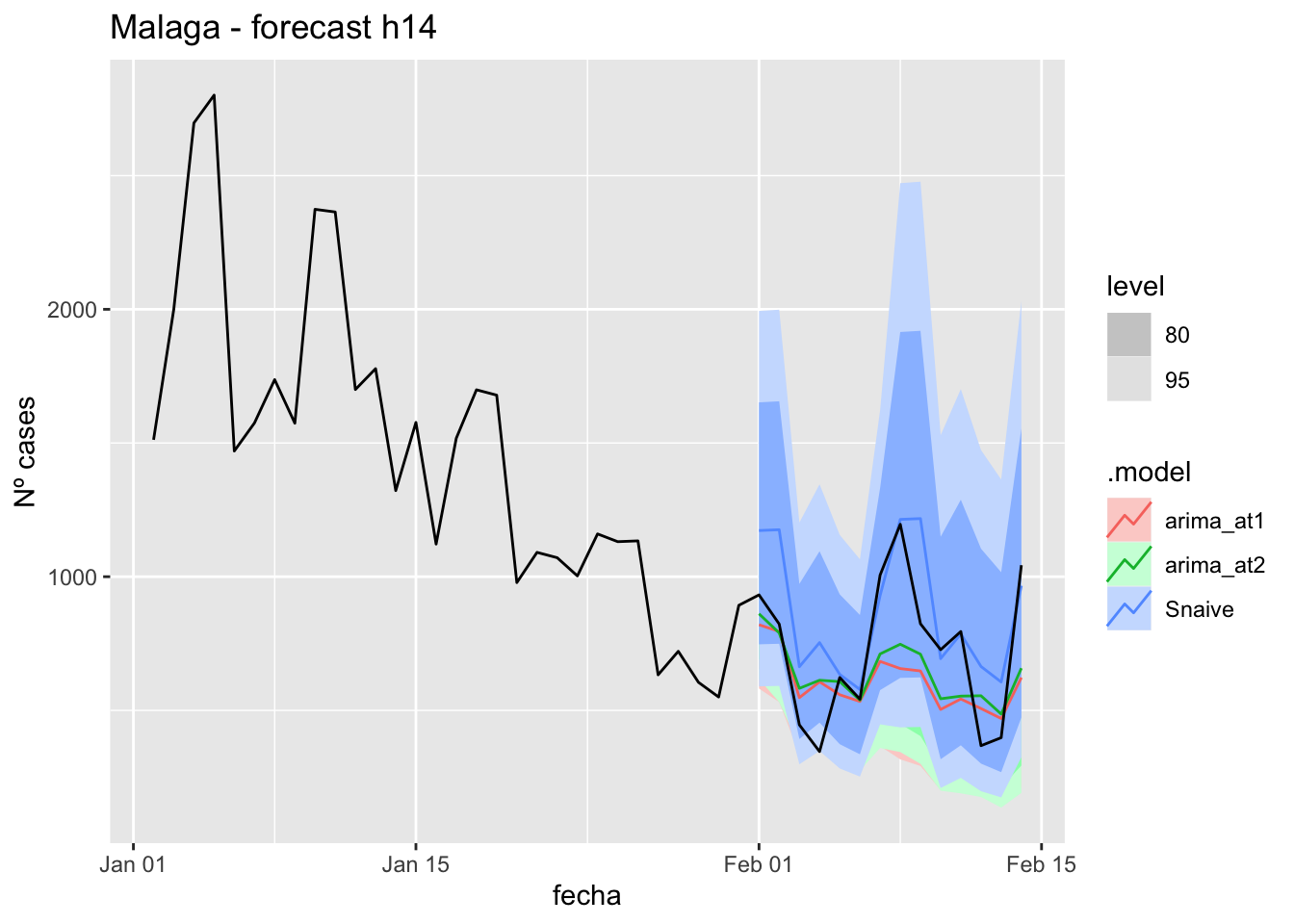
# Accuracy
fabletools::accuracy(fc_h14, data_Malaga)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_at1 Málaga Test 112. 245. 194. 5.34 27.3 1.47 0.988 0.293
2 arima_at2 Málaga Test 79.4 220. 176. 0.521 26.3 1.33 0.891 0.268
3 Snaive Málaga Test -142. 223. 170. -28.9 31.9 1.29 0.901 0.023521 days
# Plots
fc_h21 %>%
autoplot(
data_Malaga_test %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(21))
) +
labs(y = "Nº cases", title = "Malaga - forecast h21")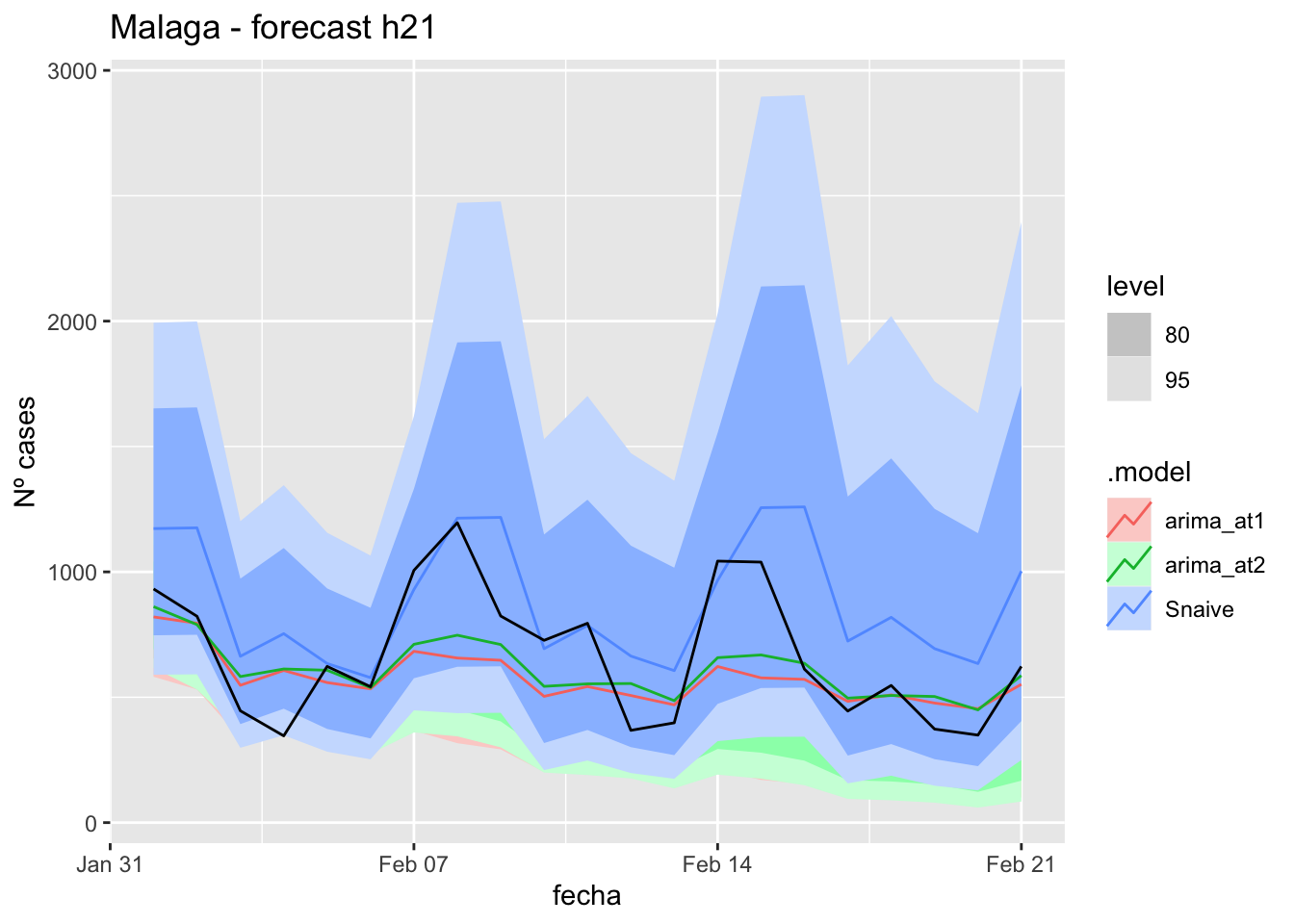
fc_h21 %>%
autoplot(
data_Malaga %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(21) &
fecha > as.Date("2022-01-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Malaga - forecast h21")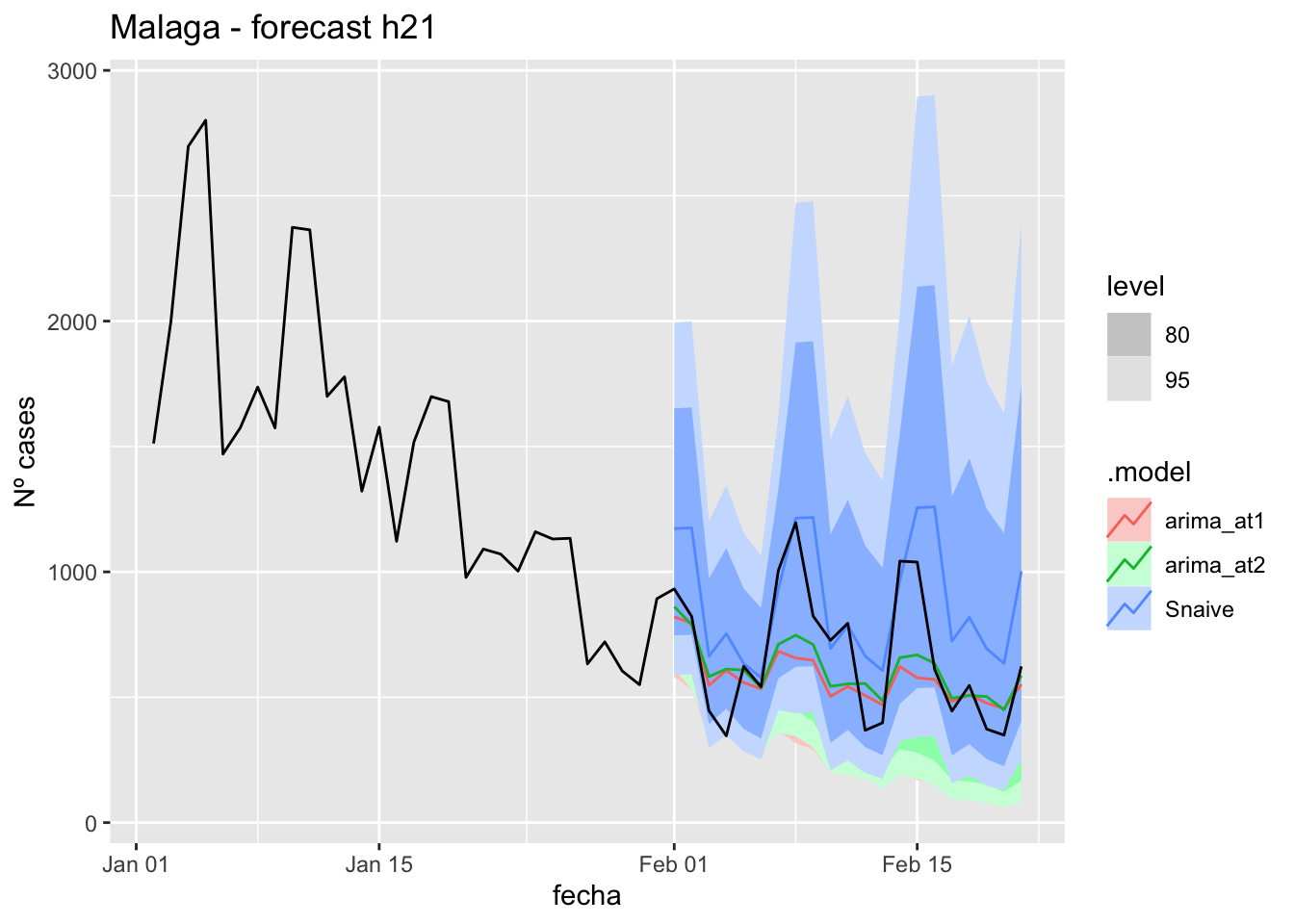
# Accuracy
fabletools::accuracy(fc_h21, data_Malaga)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_at1 Málaga Test 92.3 227. 170. 3.69 24.7 1.29 0.917 0.401
2 arima_at2 Málaga Test 59.7 201. 153. -1.09 23.6 1.16 0.813 0.364
3 Snaive Málaga Test -209. 280. 228. -41.6 43.5 1.72 1.13 0.17257 days
# Plots
fc_h57 %>%
autoplot(
data_Malaga_test %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(57))
) +
labs(y = "Nº cases", title = "Malaga - forecast h57")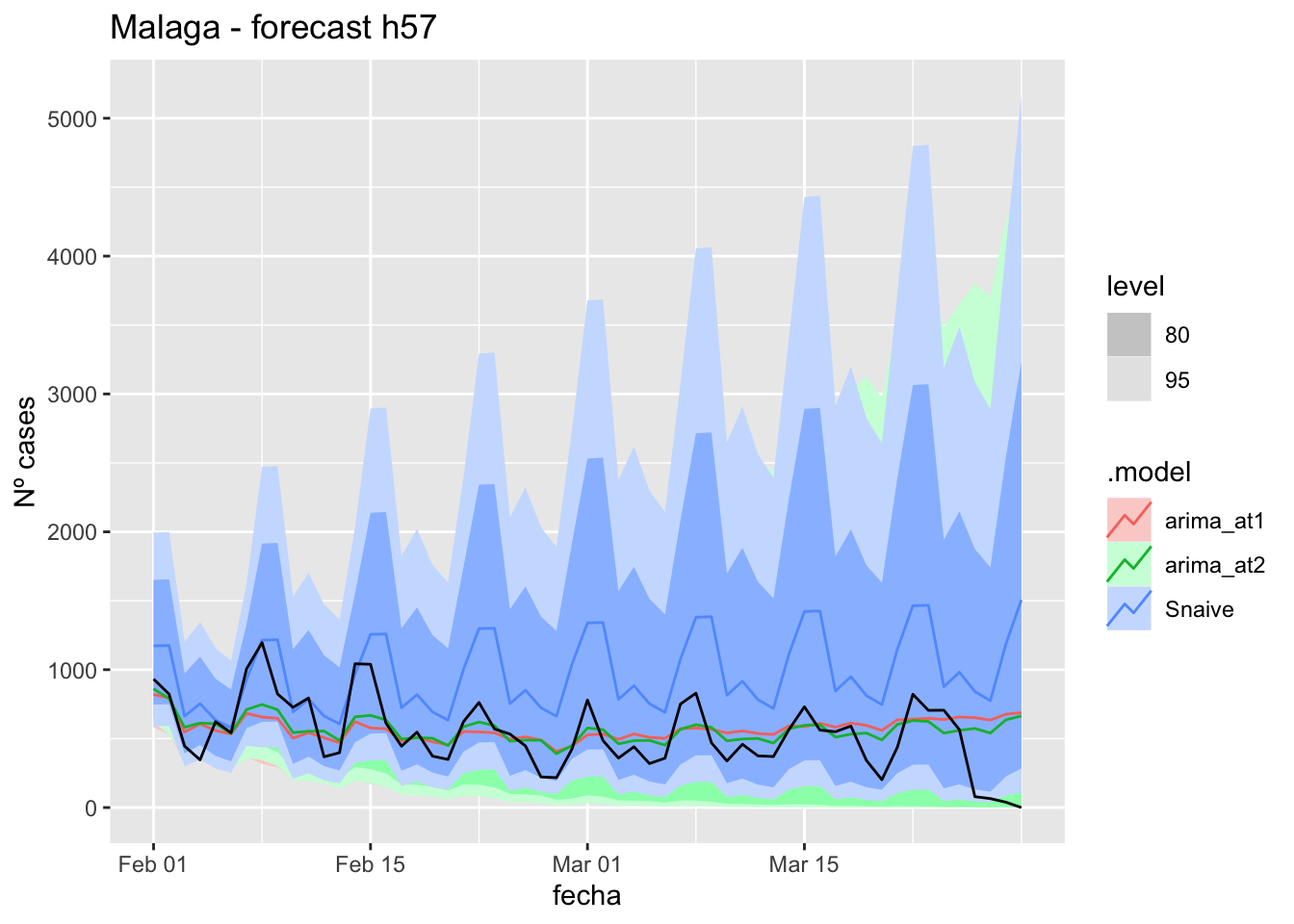
fc_h57 %>%
autoplot(
data_Malaga %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(57) &
fecha > as.Date("2022-01-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Malaga - forecast h57")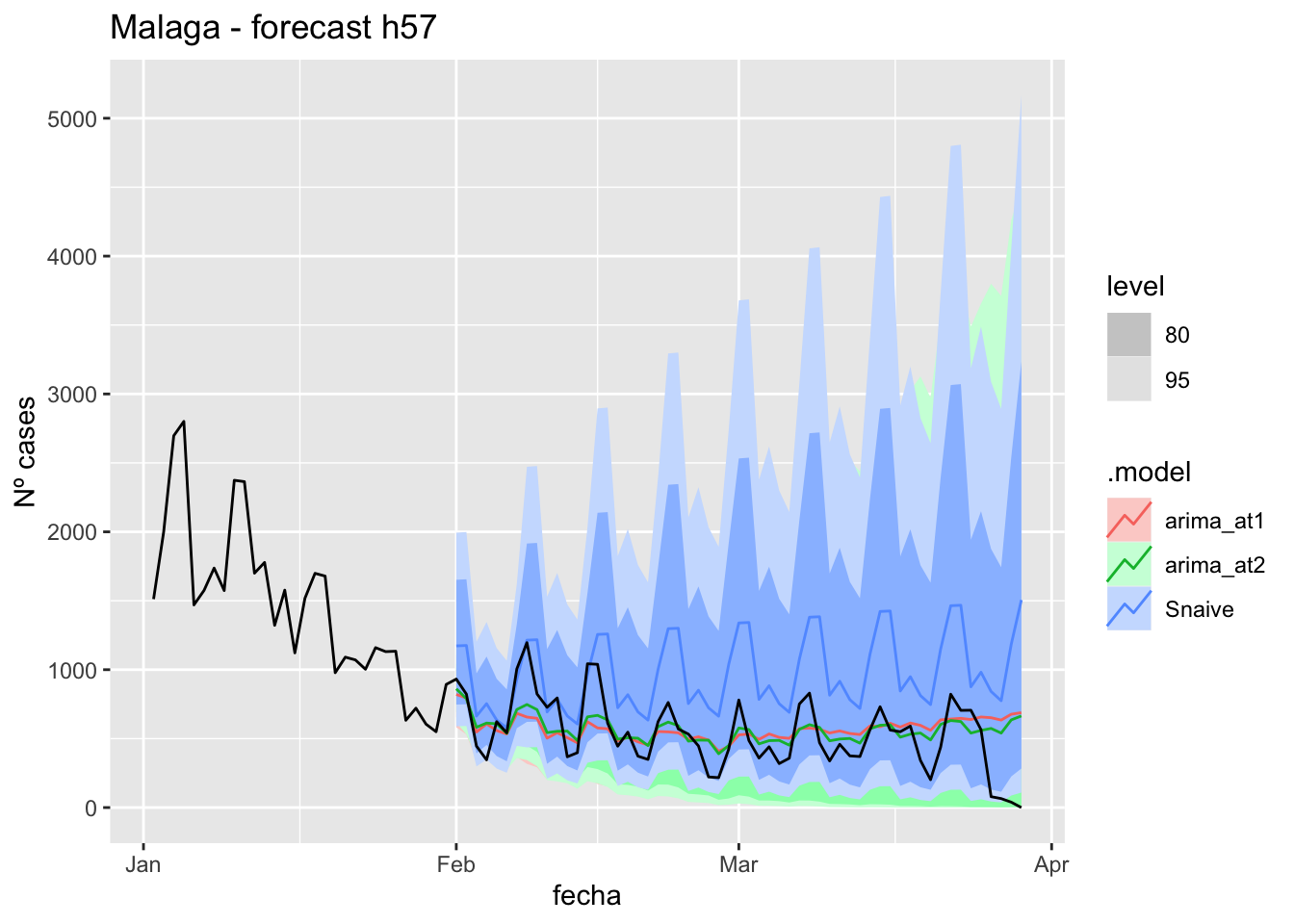
# Accuracy
fabletools::accuracy(fc_h57, data_Malaga)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_at1 Málaga Test -35.6 246. 182. -Inf Inf 1.38 0.993 0.597
2 arima_at2 Málaga Test -30.4 219. 161. -Inf Inf 1.22 0.885 0.534
3 Snaive Málaga Test -438. 527. 445. -Inf Inf 3.37 2.13 0.514Sevilla
data_Sevilla <- covid_data %>%
filter(provincia == "Sevilla") %>%
filter(fecha > as.Date("2020-06-14", format = "%Y-%m-%d")) %>%
select(provincia, fecha, num_casos, tmed, mob_grocery_pharmacy, mob_parks, mob_residential, mob_retail_recreation, mob_transit_stations, mob_workplaces) %>%
drop_na() %>%
as_tsibble(key = provincia, index = fecha)
data_Sevilla# A tsibble: 653 x 10 [1D]
# Key: provincia [1]
provincia fecha num_casos tmed mob_grocery_pharmacy mob_parks
<chr> <date> <dbl> <dbl> <dbl> <dbl>
1 Sevilla 2020-06-15 2 23.1 -14 -13
2 Sevilla 2020-06-16 1 24.0 -10 -4
3 Sevilla 2020-06-17 0 24.9 -10 -7
4 Sevilla 2020-06-18 0 25.8 -12 -13
5 Sevilla 2020-06-19 0 26.6 -13 -20
6 Sevilla 2020-06-20 0 27.5 -20 -34
7 Sevilla 2020-06-21 0 28.4 -27 -47
8 Sevilla 2020-06-22 1 29.3 -17 -19
9 Sevilla 2020-06-23 0 30.2 -12 -13
10 Sevilla 2020-06-24 1 29.4 -12 -12
# … with 643 more rows, and 4 more variables: mob_residential <dbl>,
# mob_retail_recreation <dbl>, mob_transit_stations <dbl>,
# mob_workplaces <dbl>data_Sevilla_train <- data_Sevilla %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d"))
summary(data_Sevilla_train$fecha) Min. 1st Qu. Median Mean 3rd Qu. Max.
"2020-06-15" "2020-11-10" "2021-04-08" "2021-04-08" "2021-09-04" "2022-01-31" data_Sevilla_test <- data_Sevilla %>%
filter(fecha > as.Date("2022-01-31", format = "%Y-%m-%d"))
summary(data_Sevilla_test$fecha) Min. 1st Qu. Median Mean 3rd Qu. Max.
"2022-02-01" "2022-02-15" "2022-03-01" "2022-03-01" "2022-03-15" "2022-03-29" # Lamba for num_casos
lambda_Sevilla_casos <- data_asturias %>%
features(num_casos, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Sevilla_casos[1] 0.2278229# Lamba for average temp
lambda_Sevilla_tmed <- data_asturias %>%
features(tmed, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Sevilla_tmed[1] 1.056074# Lamba for mobility
lambda_Sevilla_grocery <- data_asturias %>%
features(mob_grocery_pharmacy, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Sevilla_grocery[1] 1.377952lambda_Sevilla_parks <- data_asturias %>%
features(mob_parks, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Sevilla_parks[1] 0.7188469lambda_Sevilla_resd <- data_asturias %>%
features(mob_residential, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Sevilla_resd[1] 1.000064lambda_Sevilla_retail <- data_asturias %>%
features(mob_retail_recreation, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Sevilla_retail[1] 1.058286lambda_Sevilla_transit <- data_asturias %>%
features(mob_transit_stations, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Sevilla_transit[1] 1.116829lambda_Sevilla_work <- data_asturias %>%
features(mob_workplaces, features = guerrero) %>%
pull(lambda_guerrero)
lambda_Sevilla_work[1] 1.999927fit_model <- data_Sevilla_train %>%
model(
arima_at1 = ARIMA(box_cox(num_casos, lambda_Sevilla_casos) ~
box_cox(tmed, lambda_Sevilla_tmed) +
box_cox(mob_grocery_pharmacy, lambda_Sevilla_grocery) +
box_cox(mob_parks, lambda_Sevilla_parks) +
box_cox(mob_residential, lambda_Sevilla_resd) +
box_cox(mob_retail_recreation, lambda_Sevilla_retail) +
box_cox(mob_transit_stations, lambda_Sevilla_transit) +
box_cox(mob_workplaces, lambda_Sevilla_work)
),
arima_at2 = ARIMA(box_cox(num_casos, lambda_Sevilla_casos) ~
box_cox(tmed, lambda_Sevilla_tmed) +
box_cox(mob_grocery_pharmacy, lambda_Sevilla_grocery) +
box_cox(mob_parks, lambda_Sevilla_parks) +
box_cox(mob_residential, lambda_Sevilla_resd) +
box_cox(mob_retail_recreation, lambda_Sevilla_retail) +
box_cox(mob_transit_stations, lambda_Sevilla_transit) +
box_cox(mob_workplaces, lambda_Sevilla_work),
stepwise = FALSE, approx = FALSE),
Snaive = SNAIVE(box_cox(num_casos, lambda_Sevilla_casos))
)
# Show and report model
fit_model# A mable: 1 x 4
# Key: provincia [1]
provincia arima_at1
<chr> <model>
1 Sevilla <LM w/ ARIMA(3,1,1)(2,0,0)[7] errors>
# … with 2 more variables: arima_at2 <model>, Snaive <model>glance(fit_model)# A tibble: 3 × 9
provincia .model sigma2 log_lik AIC AICc BIC ar_roots ma_roots
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <list> <list>
1 Sevilla arima_at1 0.940 -821. 1670. 1670. 1731. <cpl [17]> <cpl [1]>
2 Sevilla arima_at2 0.910 -813. 1654. 1654. 1715. <cpl [7]> <cpl [11]>
3 Sevilla Snaive 2.93 NA NA NA NA <NULL> <NULL> fit_model %>%
pivot_longer(-provincia,
names_to = ".model",
values_to = "orders") %>%
left_join(glance(fit_model), by = c("provincia", ".model")) %>%
arrange(AICc) %>%
select(.model:BIC)# A mable: 3 x 7
# Key: .model [3]
.model orders sigma2 log_lik AIC AICc BIC
<chr> <model> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_… <LM w/ ARIMA(0,1,4)(1,0,1)[7] errors> 0.910 -813. 1654. 1654. 1715.
2 arima_… <LM w/ ARIMA(3,1,1)(2,0,0)[7] errors> 0.940 -821. 1670. 1670. 1731.
3 Snaive <SNAIVE> 2.93 NA NA NA NA # We use a Ljung-Box test >> large p-value, confirms residuals are similar / considered to white noise
fit_model %>%
select(Snaive) %>%
gg_tsresiduals()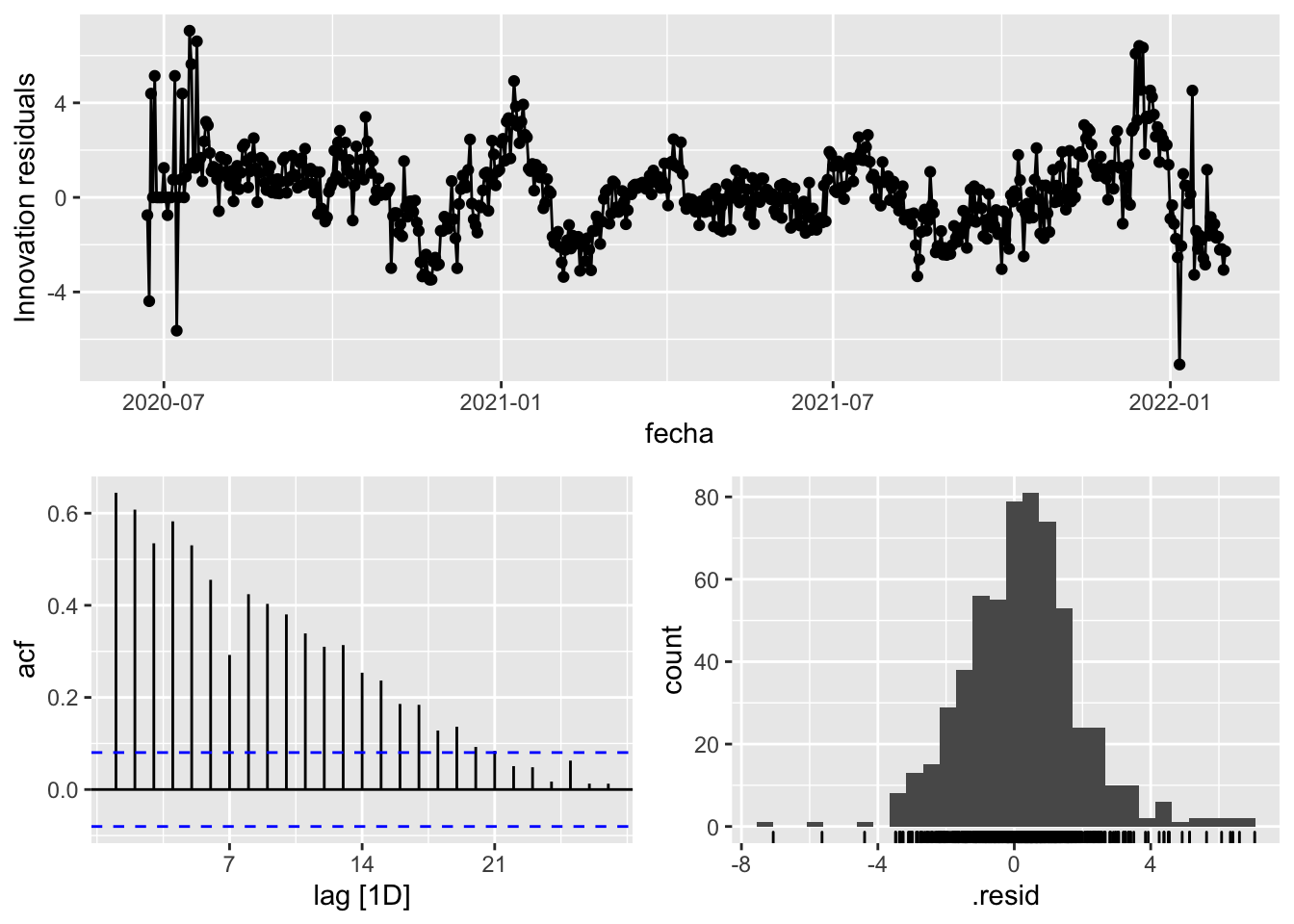
fit_model %>%
select(arima_at1) %>%
gg_tsresiduals()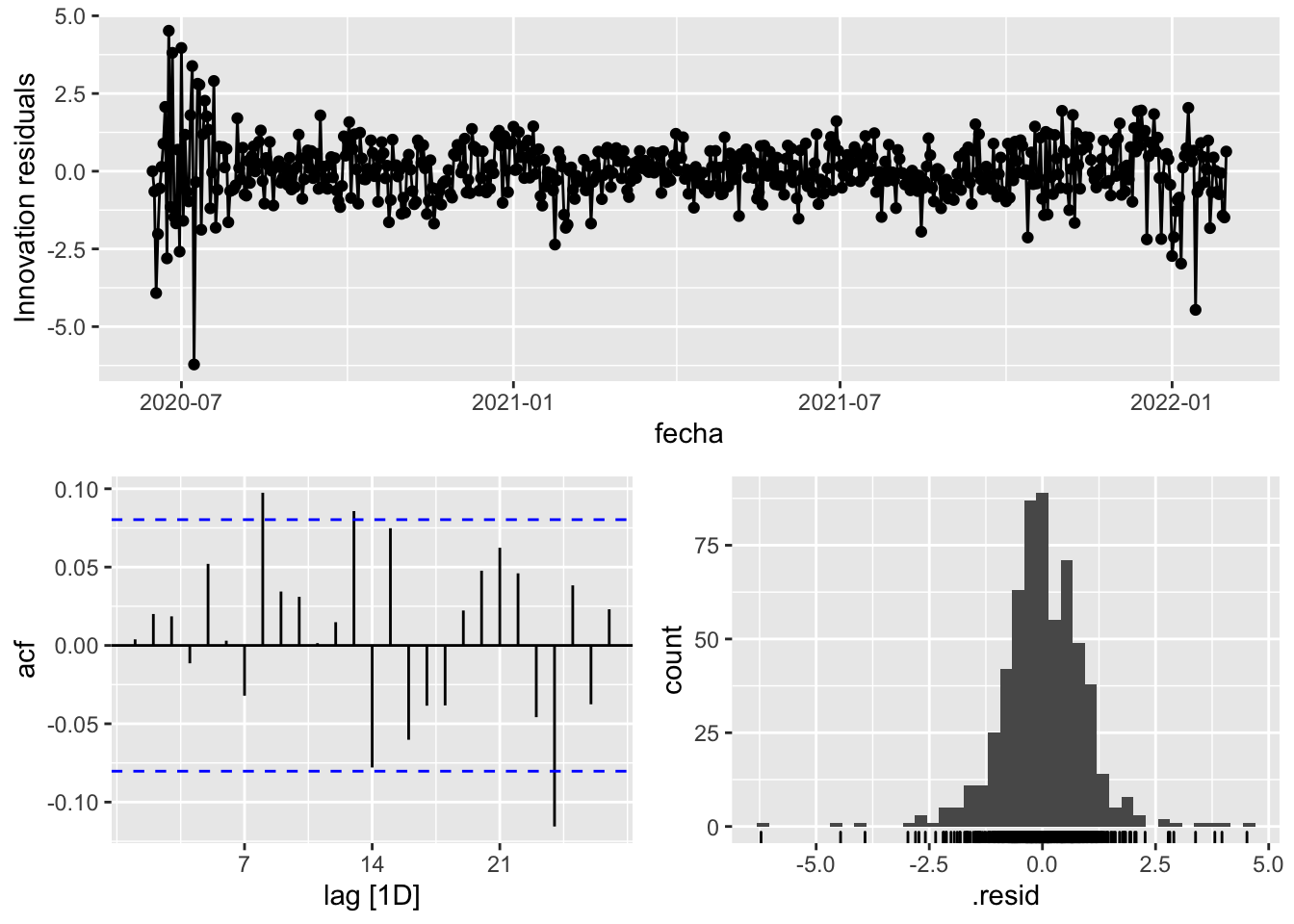
fit_model %>%
select(arima_at2) %>%
gg_tsresiduals()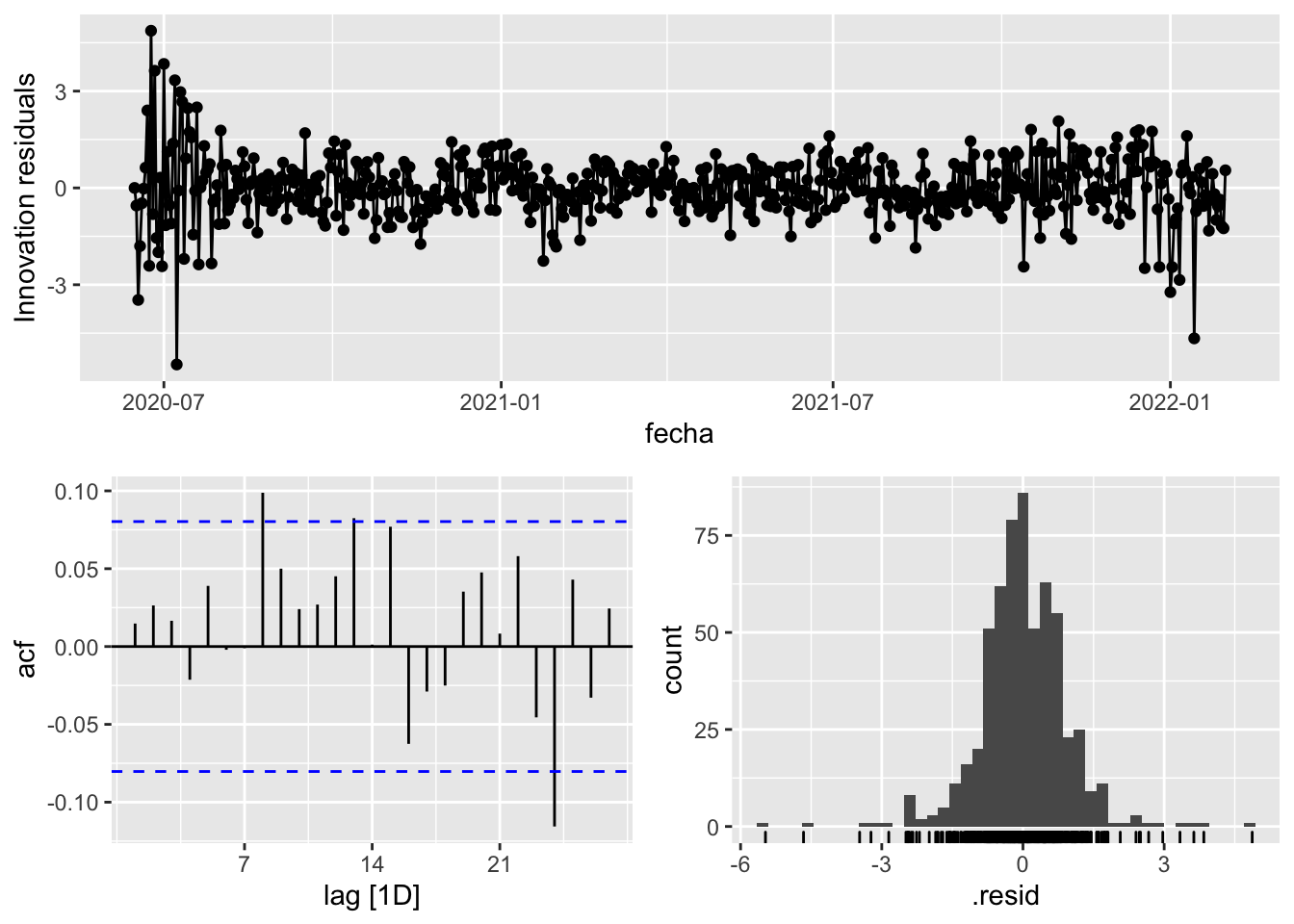
augment(fit_model) %>%
features(.innov, ljung_box, lag=7)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Sevilla arima_at1 2.80 0.903
2 Sevilla arima_at2 1.90 0.965
3 Sevilla Snaive 1179. 0 augment(fit_model) %>%
features(.innov, ljung_box, lag=14)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Sevilla arima_at1 18.2 0.198
2 Sevilla arima_at2 15.5 0.343
3 Sevilla Snaive 1697. 0 augment(fit_model) %>%
features(.innov, ljung_box, lag=21)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Sevilla arima_at1 29.8 0.0960
2 Sevilla arima_at2 24.7 0.261
3 Sevilla Snaive 1803. 0 augment(fit_model) %>%
features(.innov, ljung_box, lag=57)# A tibble: 3 × 4
provincia .model lb_stat lb_pvalue
<chr> <chr> <dbl> <dbl>
1 Sevilla arima_at1 74.5 0.0599
2 Sevilla arima_at2 71.2 0.0979
3 Sevilla Snaive 2248. 0 # To predict future values of infections, we need to pass the model future/prediction values of the complementary variables tmed and mobility
# We are going to select those from the test dataset
data_fc_h7 <- data_Sevilla_test %>%
select(-num_casos) %>%
slice(1:7)
data_fc_h14 <- data_Sevilla_test %>%
select(-num_casos) %>%
slice(1:14)
data_fc_h21 <- data_Sevilla_test %>%
select(-num_casos) %>%
slice(1:21)
data_fc_h57 <- data_Sevilla_test %>%
select(-num_casos) %>%
slice(1:57)# Significant spikes out of 30 is still consistent with white noise
# To be sure, use a Ljung-Box test, which has a large p-value, confirming that
# the residuals are similar to white noise.
# Note that the alternative models also passed this test.
#
# Forecast
fc_h7 <- fabletools::forecast(fit_model, new_data = data_fc_h7)
fc_h14 <- fabletools::forecast(fit_model, new_data = data_fc_h14)
fc_h21 <- fabletools::forecast(fit_model, new_data = data_fc_h21)
fc_h57 <- fabletools::forecast(fit_model, new_data = data_fc_h57)7 days
# Plots
fc_h7 %>%
autoplot(
data_Sevilla_test %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(7))
) +
labs(y = "Nº cases", title = "Sevilla - forecast h7")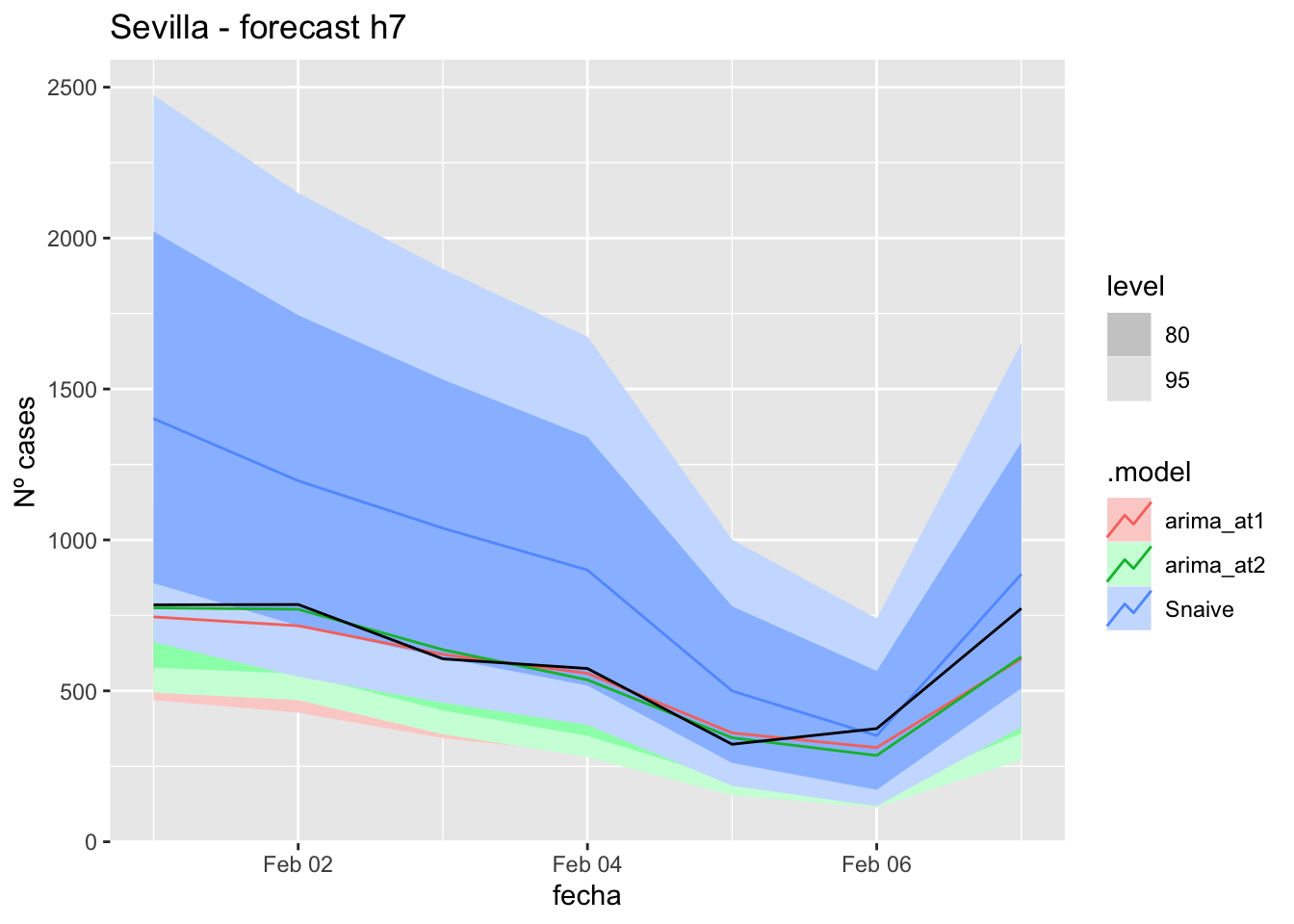
fc_h7 %>%
autoplot(
data_Sevilla %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(7) &
fecha > as.Date("2022-01-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Sevilla - forecast h7")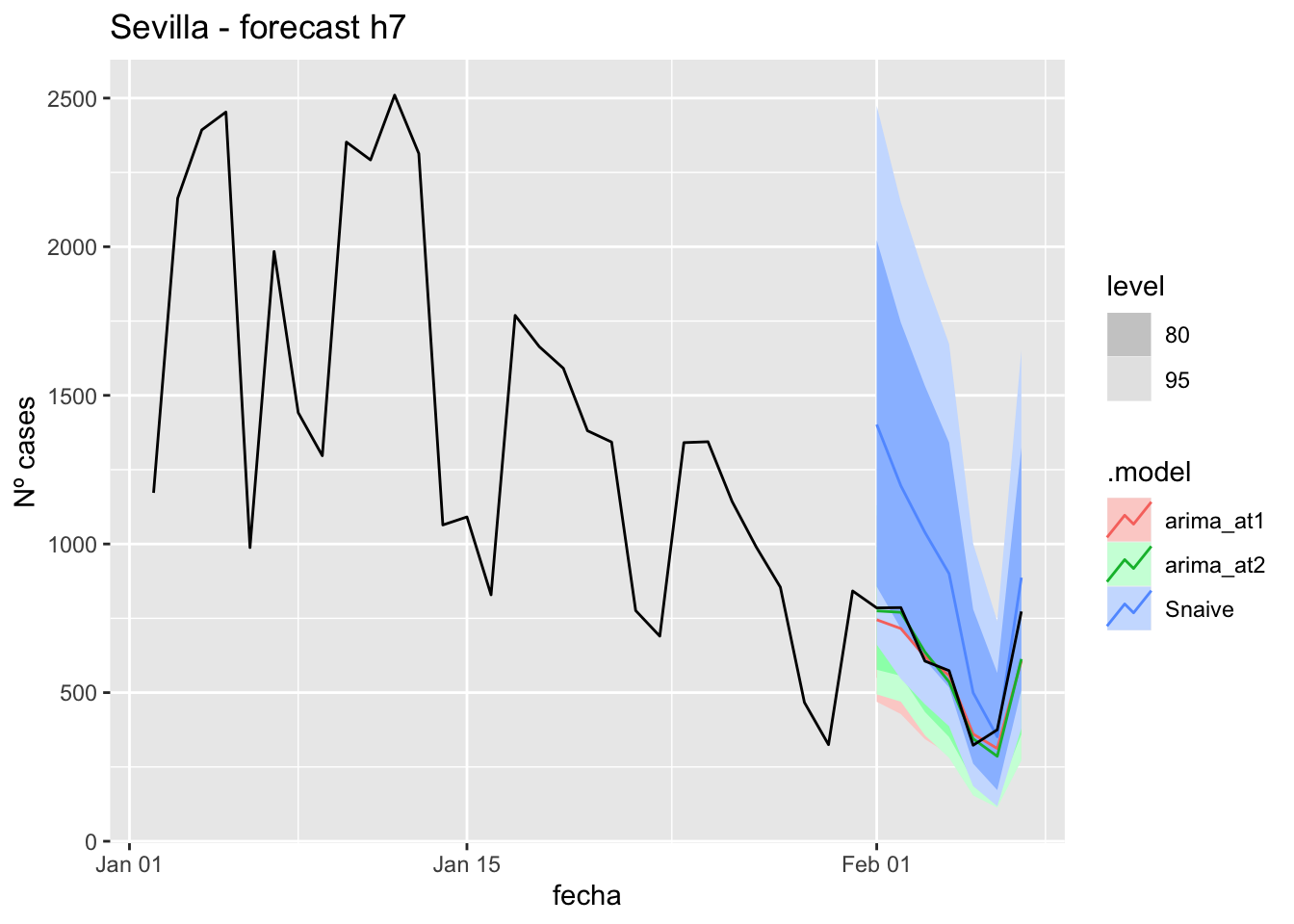
# Accuracy
fabletools::accuracy(fc_h7, data_Sevilla)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_at1 Sevilla Test 43.3 75.6 58.3 5.86 9.89 0.407 0.277 0.110
2 arima_at2 Sevilla Test 37.2 72.4 52.1 6.09 9.43 0.364 0.265 0.196
3 Snaive Sevilla Test -293. 356. 300. -46.0 47.8 2.10 1.30 0.52114 days
# Plots
fc_h14 %>%
autoplot(
data_Sevilla_test %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(14))
) +
labs(y = "Nº cases", title = "Sevilla - forecast h14")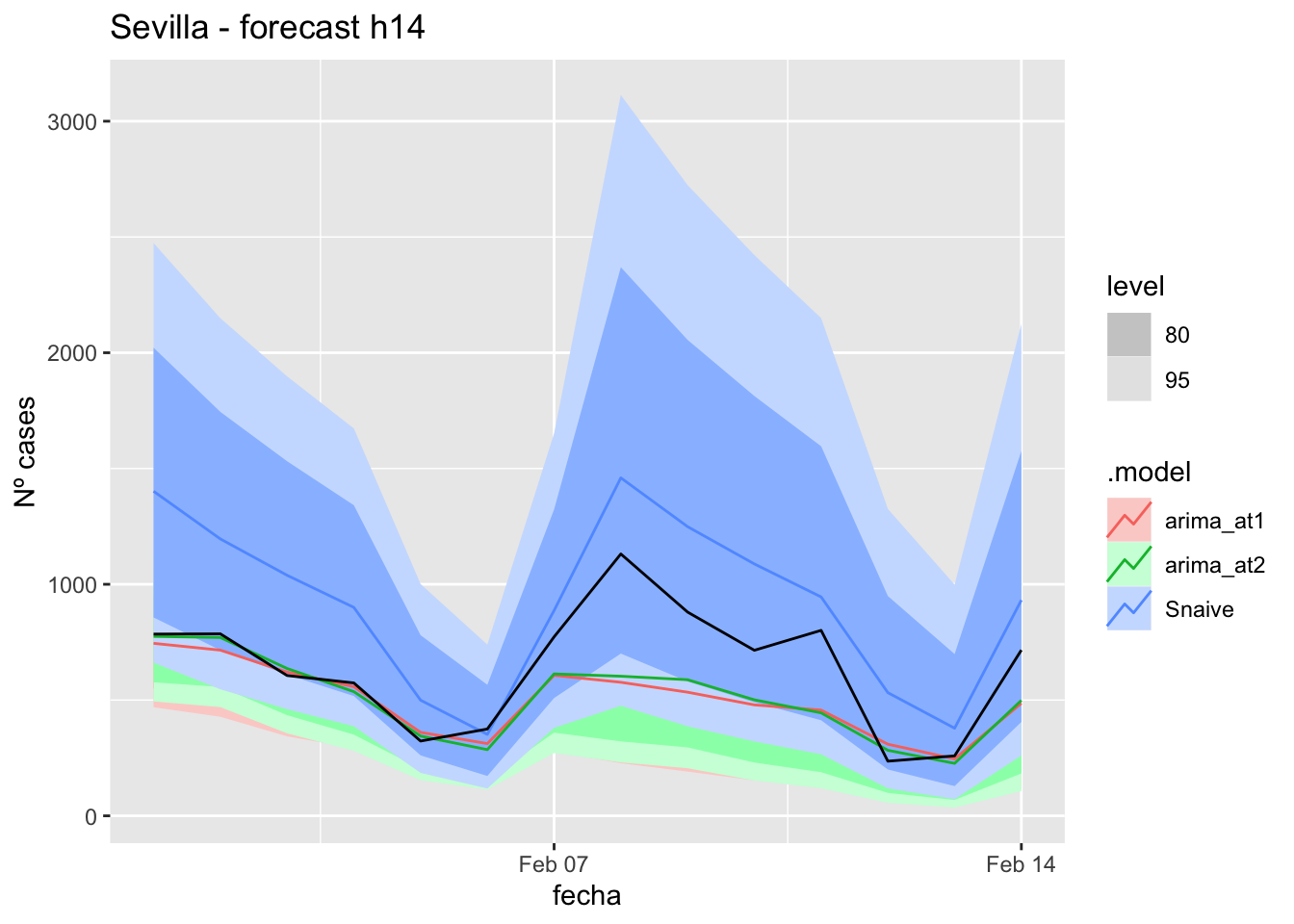
fc_h14 %>%
autoplot(
data_Sevilla %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(14) &
fecha > as.Date("2022-01-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Sevilla - forecast h14")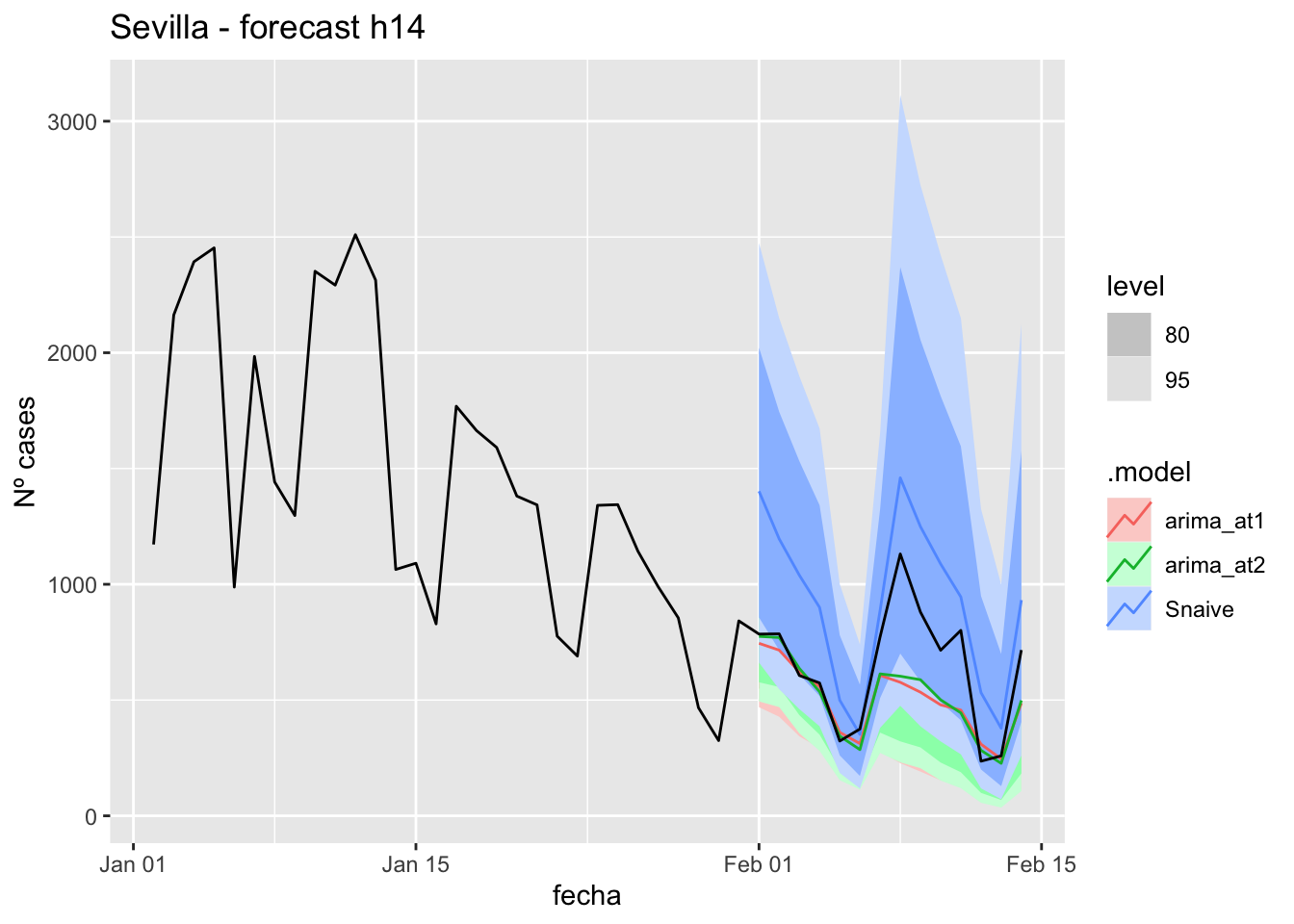
# Accuracy
fabletools::accuracy(fc_h14, data_Sevilla)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_at1 Sevilla Test 139. 224. 157. 15.1 21.6 1.10 0.819 0.417
2 arima_at2 Sevilla Test 132. 211. 147. 15.7 20.2 1.02 0.773 0.379
3 Snaive Sevilla Test -279. 321. 282. -47.5 48.4 1.97 1.17 0.40921 days
# Plots
fc_h21 %>%
autoplot(
data_Sevilla_test %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(21))
) +
labs(y = "Nº cases", title = "Sevilla - forecast h21")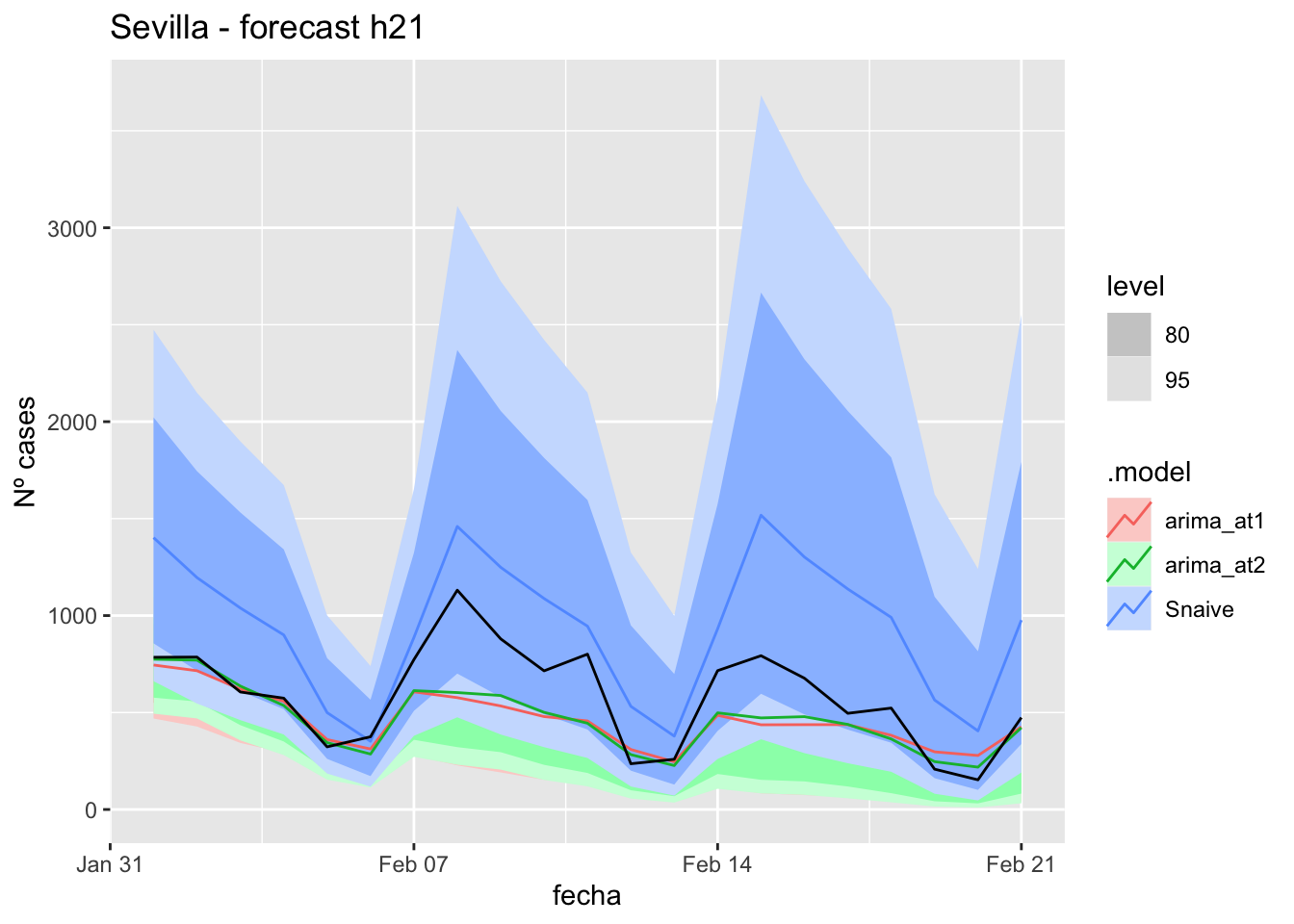
fc_h21 %>%
autoplot(
data_Sevilla %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(21) &
fecha > as.Date("2022-01-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Sevilla - forecast h21")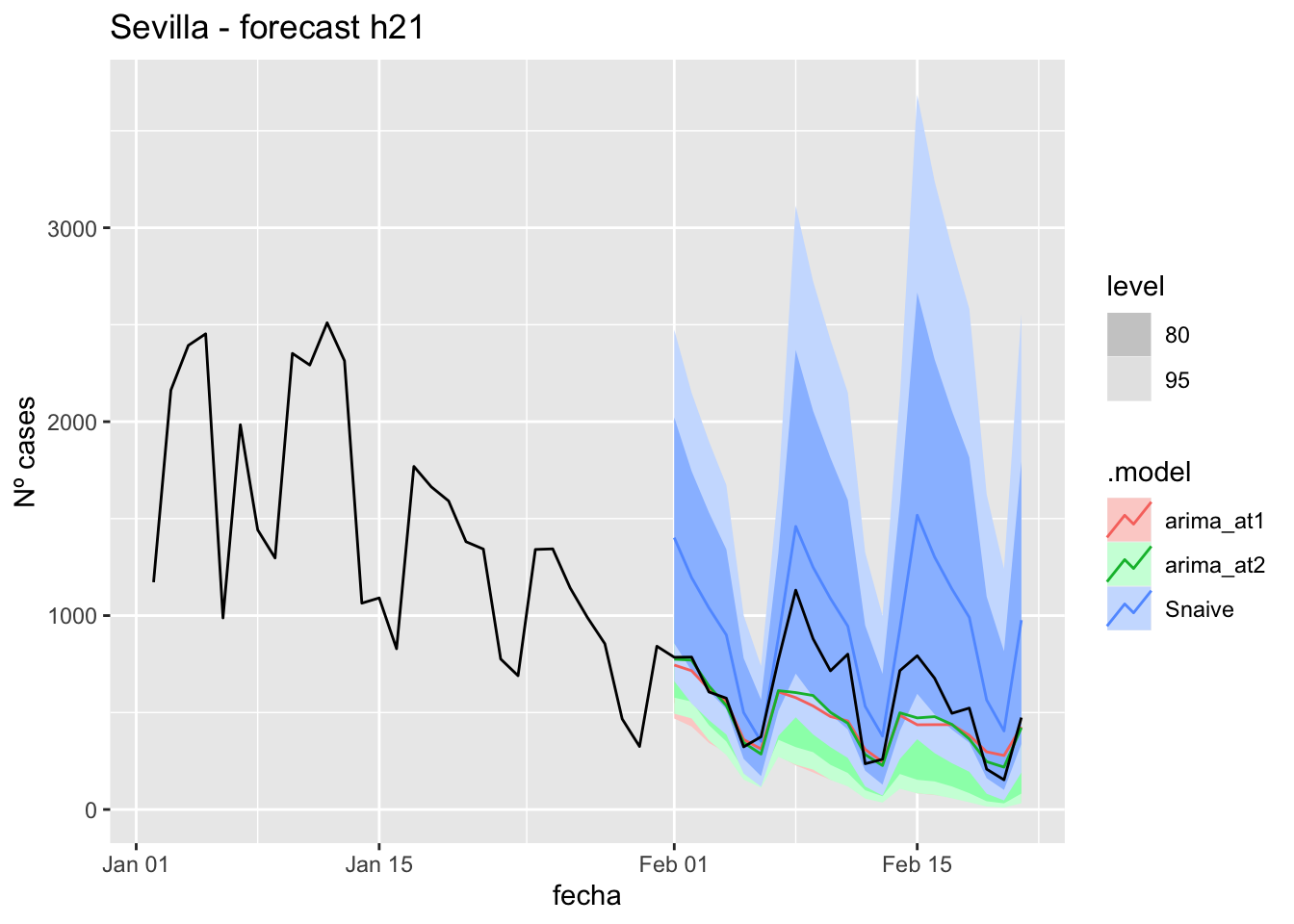
# Accuracy
fabletools::accuracy(fc_h21, data_Sevilla)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_at1 Sevilla Test 123. 211. 155. 10.3 26.5 1.09 0.772 0.476
2 arima_at2 Sevilla Test 121. 196. 140. 13.4 22.3 0.980 0.716 0.421
3 Snaive Sevilla Test -356. 404. 358. -71.9 72.5 2.50 1.48 0.49857 days
# Plots
fc_h57 %>%
autoplot(
data_Sevilla_test %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(57))
) +
labs(y = "Nº cases", title = "Sevilla - forecast h57")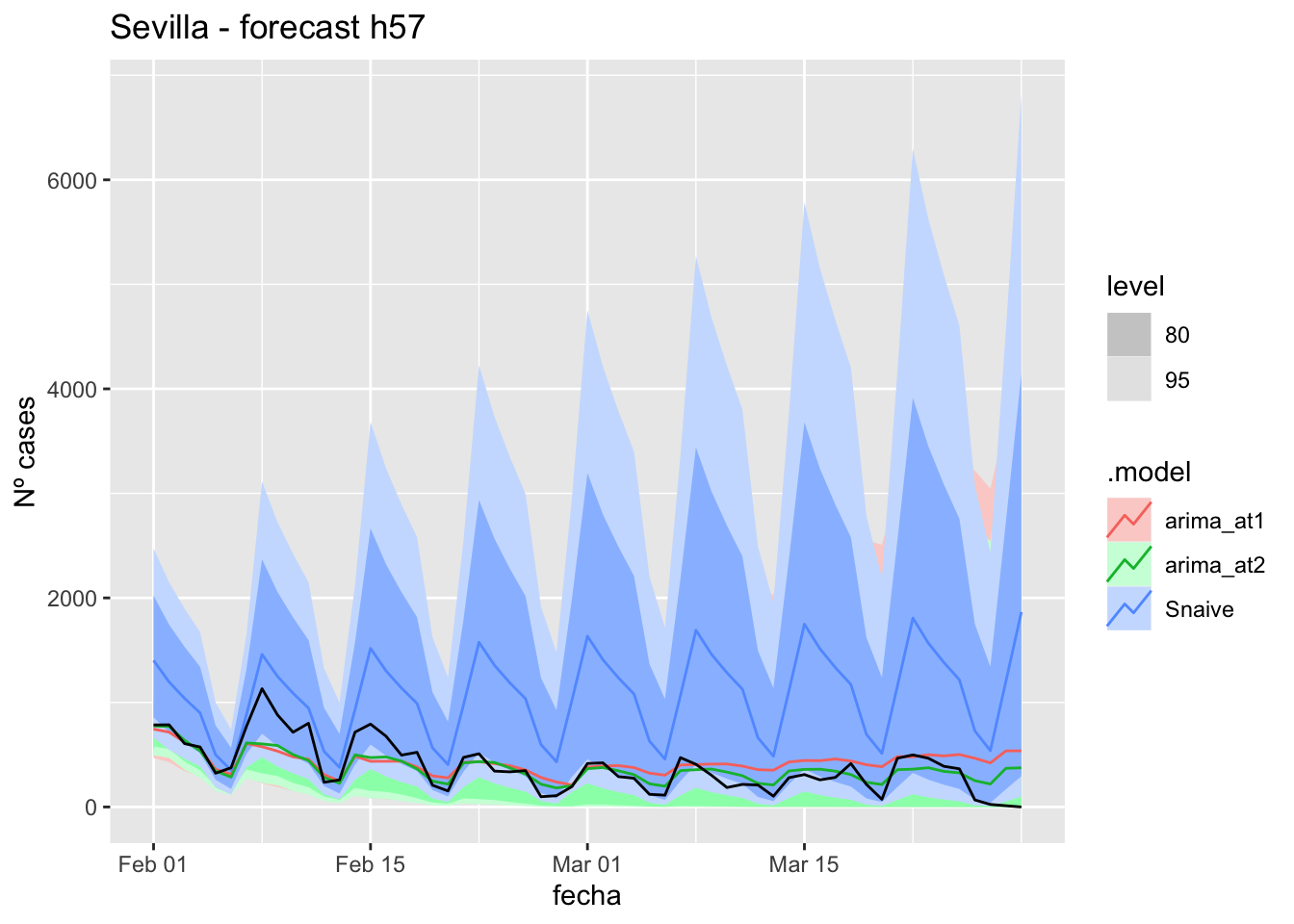
fc_h57 %>%
autoplot(
data_Sevilla %>%
filter(fecha <= as.Date("2022-01-31", format = "%Y-%m-%d")+days(57) &
fecha > as.Date("2022-01-01", format = "%Y-%m-%d"))
) +
labs(y = "Nº cases", title = "Sevilla - forecast h57")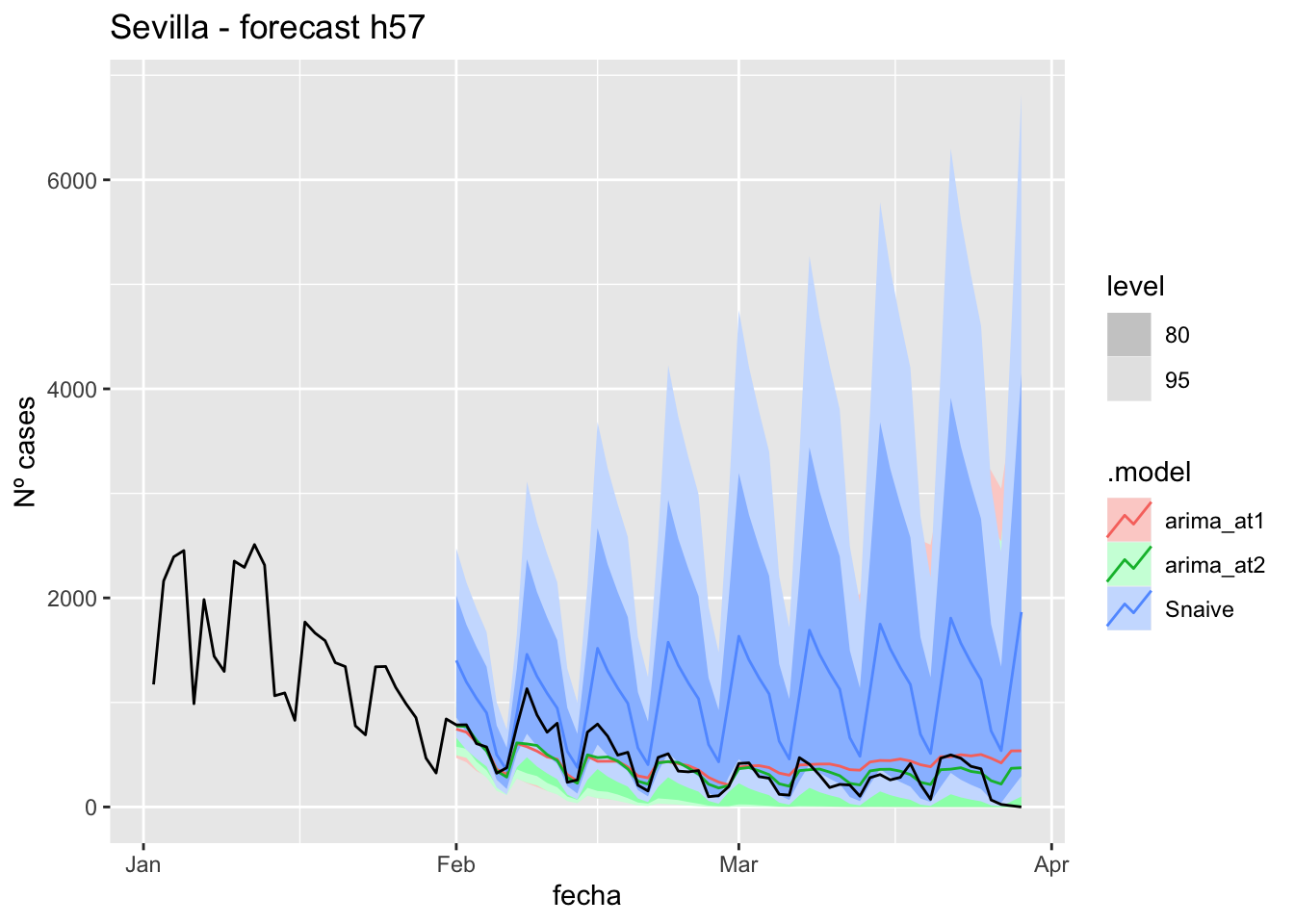
# Accuracy
fabletools::accuracy(fc_h57, data_Sevilla)# A tibble: 3 × 11
.model provincia .type ME RMSE MAE MPE MAPE MASE RMSSE ACF1
<chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 arima_at1 Sevilla Test -43.3 207. 154. -Inf Inf 1.08 0.756 0.707
2 arima_at2 Sevilla Test 15.5 155. 113. -Inf Inf 0.786 0.566 0.597
3 Snaive Sevilla Test -681. 784. 682. -Inf Inf 4.77 2.87 0.637